Search Count: 237
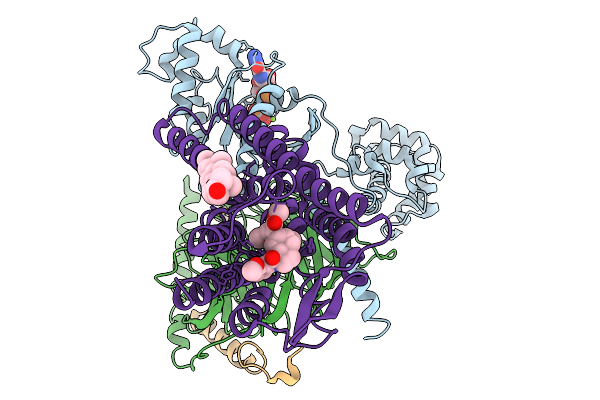 |
Structure Of The Mor/Gi/Mitragynine Pseudoindoxil Complex, Gtp-Bound G-Primed, Ahd 3Dva Sorted
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: MEMBRANE PROTEIN Ligands: CLR, EIG, GTP, MG |
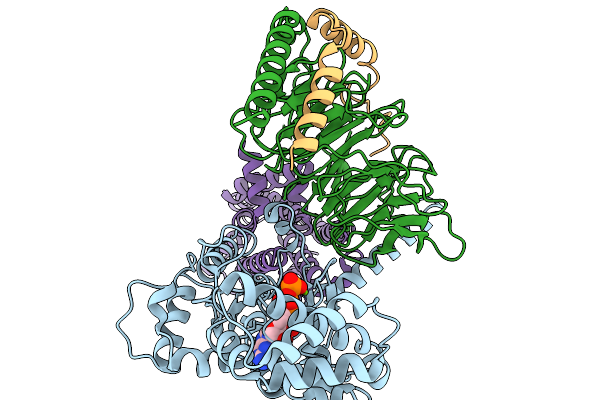 |
Structure Of The Mor/Gi/Mitragynine Pseudoindoxil Complex, Gtp-Bound G-Act-2
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: MEMBRANE PROTEIN Ligands: MG, GTP |
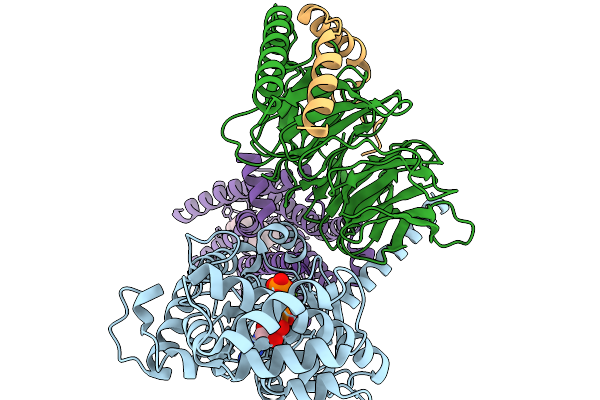 |
Structure Of The Mor/Gi/Mitragynine Pseudoindoxil Complex, Gtp-Bound G-Act-3
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: MEMBRANE PROTEIN Ligands: MG, GTP, EIG |
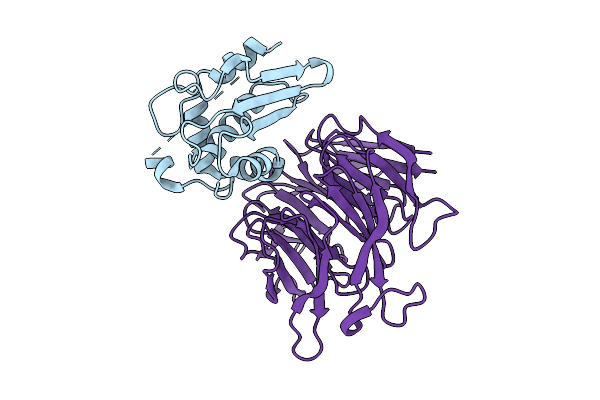 |
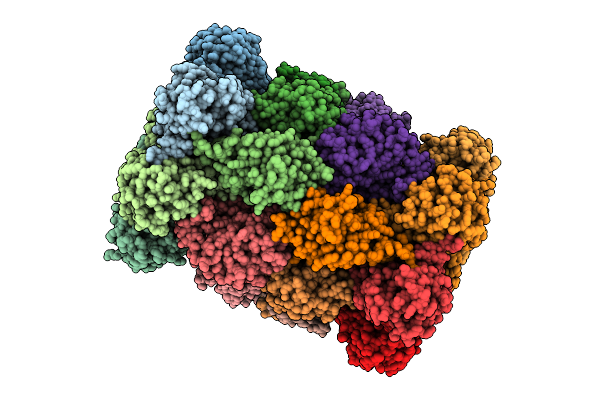
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: METAL BINDING PROTEIN |
 |
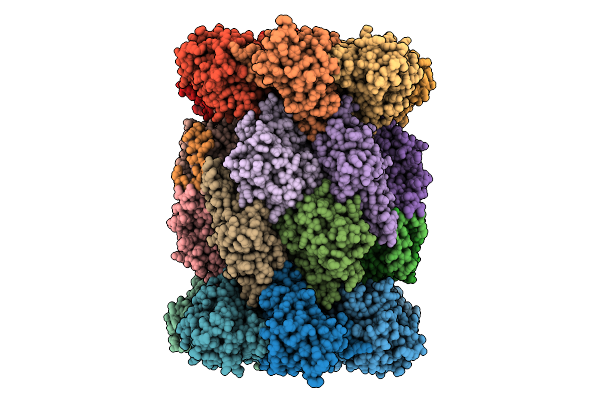
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: CYTOSOLIC PROTEIN Ligands: A1B74 |
 |
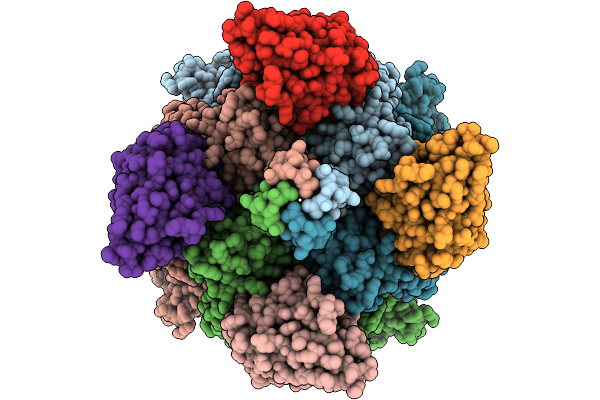
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: CYTOSOLIC PROTEIN Ligands: A1B73 |
 |
Organism: Camelus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: METAL TRANSPORT Ligands: ZN, CA, A1L5I |
 |
Organism: Homo sapiens, Camelus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: METAL TRANSPORT Ligands: ZN, CA, A1L5I |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: METAL TRANSPORT Ligands: ZN, A1L5P |
 |
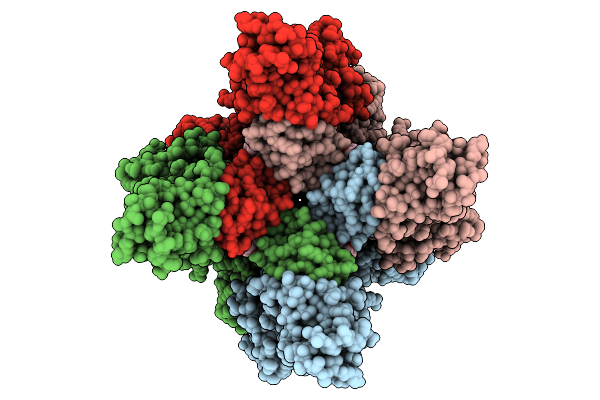
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: METAL TRANSPORT Ligands: ZN, A1L5Q |
 |
Organism: Homo sapiens, Camelus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: METAL TRANSPORT Ligands: CA, ZN, A1L5I |
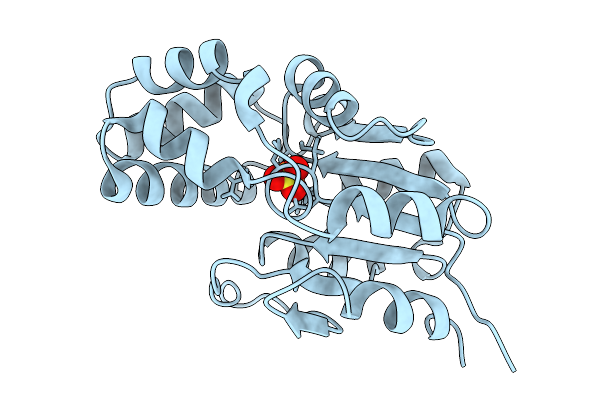 |
Crystal Structure Of Fluoroacetate Dehalogenase 4A From Delftia Acidovorans Strain D4B
Organism: Delftia acidovorans
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: TRANSFERASE Ligands: SO4, CL |
 |
Crystal Structures Fluoroacetate Dehalogenase D4B From Delftia Acidovorans Strain D4B
Organism: Delftia acidovorans
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: TRANSFERASE |
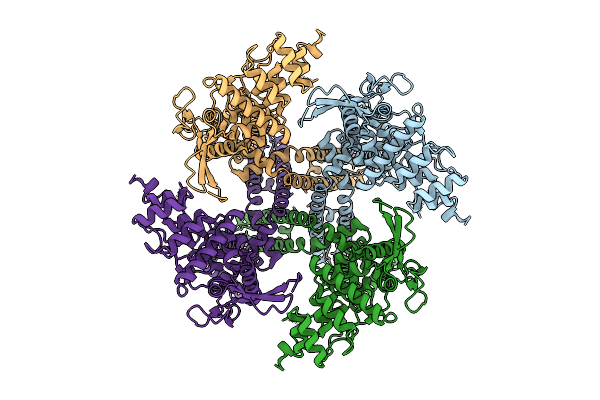 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: A1D92 |
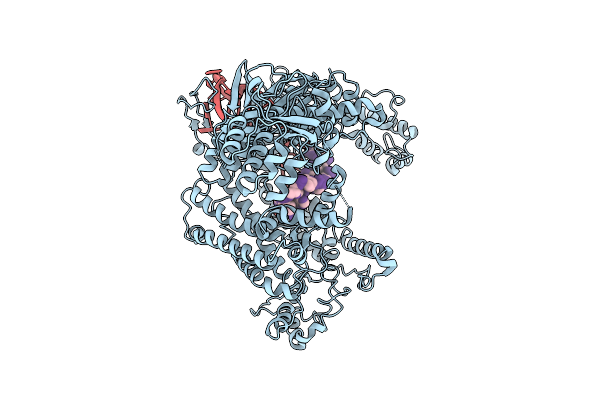 |
Organism: Lachnospiraceae bacterium nd2006, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: DNA BINDING PROTEIN/RNA |
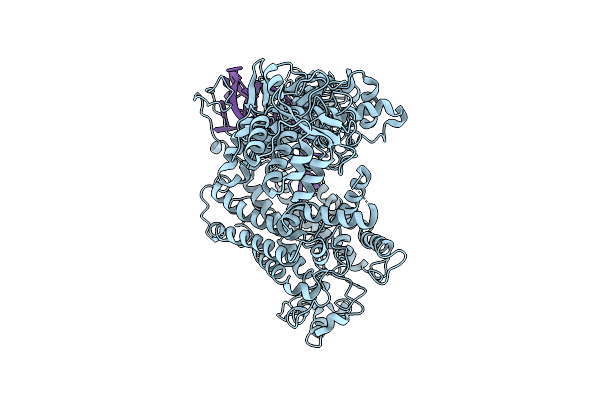 |
Organism: Lachnospiraceae bacterium nd2006, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: header DNA BINDING PROTEIN/RNA |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: PROTEIN FIBRIL |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: TRANSFERASE Ligands: COA |
 |
Organism: Orthotospovirus tomatomaculae
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: VIRAL PROTEIN Ligands: RTP |

