Search Count: 529
 |
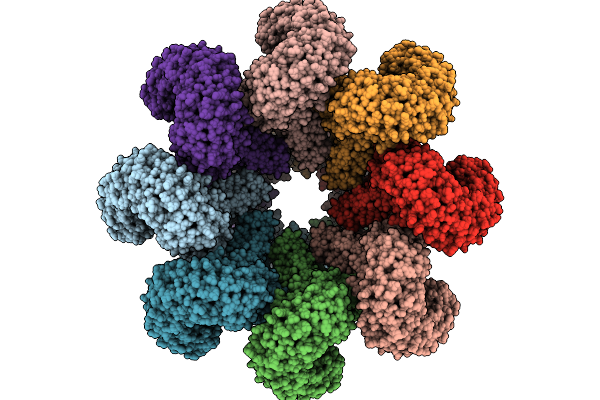 |
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PLANT PROTEIN Ligands: ATP |
 |
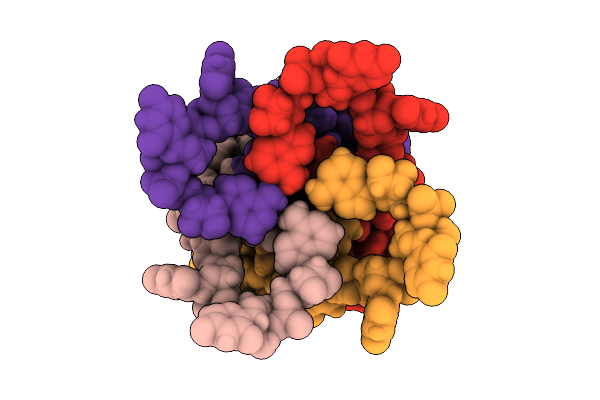
Cryo-Em Structure Of An Octameric G10-Resistosome From Wheat (N-To-N Arrangement)
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PLANT PROTEIN Ligands: ATP |
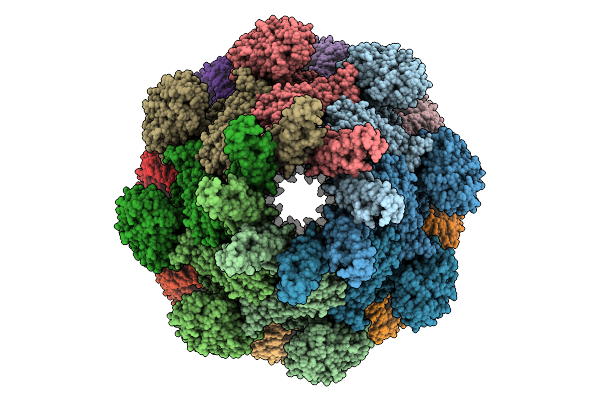 |
Cryo-Em Structure Of An Octameric G10-Resistosome From Wheat In 'Back-To-Back' Arrangement
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PLANT PROTEIN Ligands: ATP |
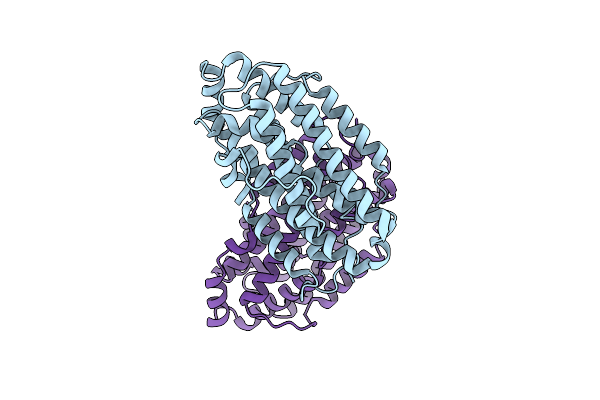 |
X-Ray Crystal Structure Of Francisella Hispaniensis Apo Ribonucleotide Reductase Beta Subunit
Organism: Francisella hispaniensis
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: OXIDOREDUCTASE Ligands: CL |
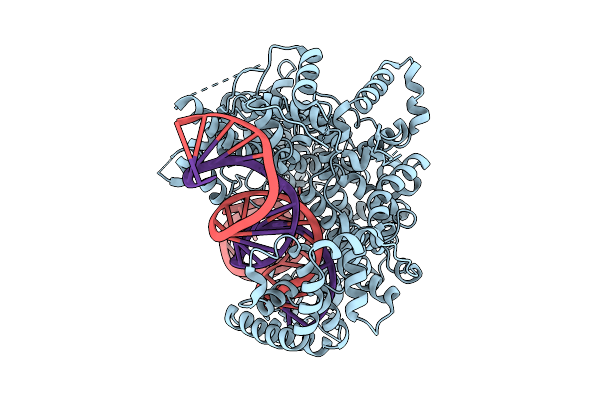 |
Organism: [eubacterium] siraeum dsm 15702
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: RNA BINDING PROTEIN/RNA Ligands: MG |
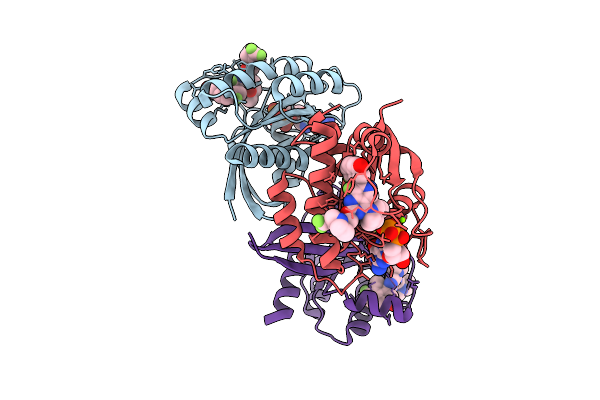 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ONCOPROTEIN Ligands: MG, A1EN3, GDP |
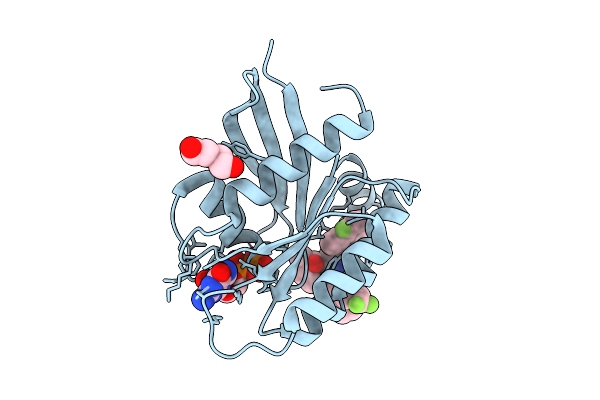 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ONCOPROTEIN Ligands: MG, A1EN3, GDP, PEG |
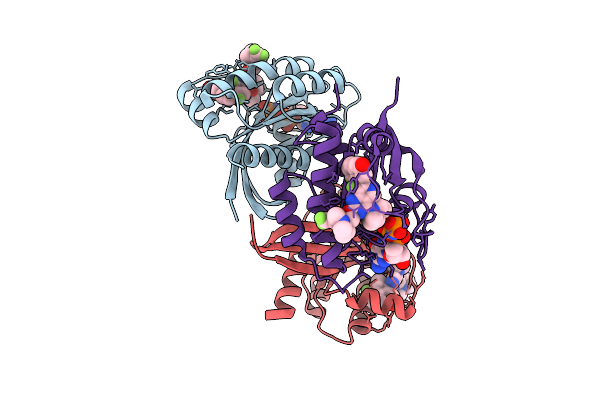 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ONCOPROTEIN Ligands: MG, A1EN3, GNP |
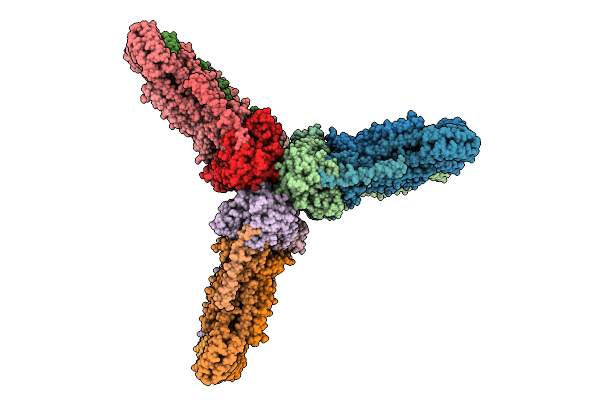 |
Organism: Escherichia coli str. k-12 substr. mg1655
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: CELL CYCLE Ligands: AGS |
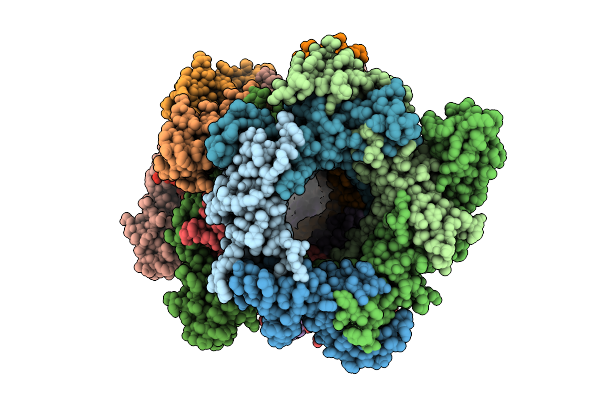 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: ANTIVIRAL PROTEIN |
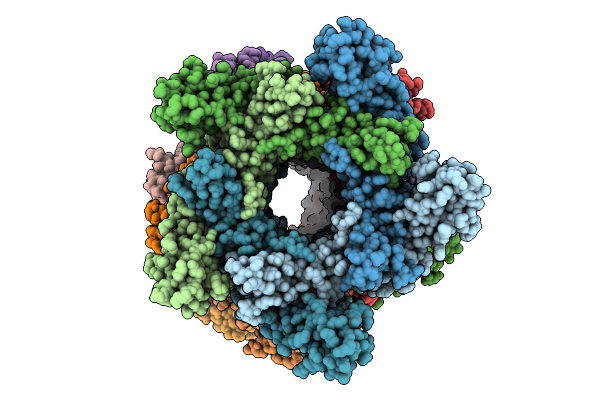 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: ANTIVIRAL PROTEIN |
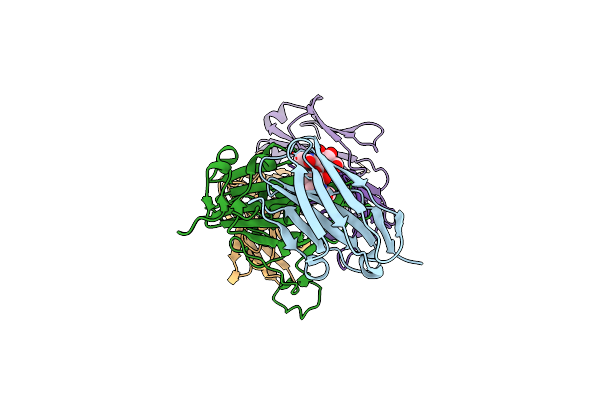 |
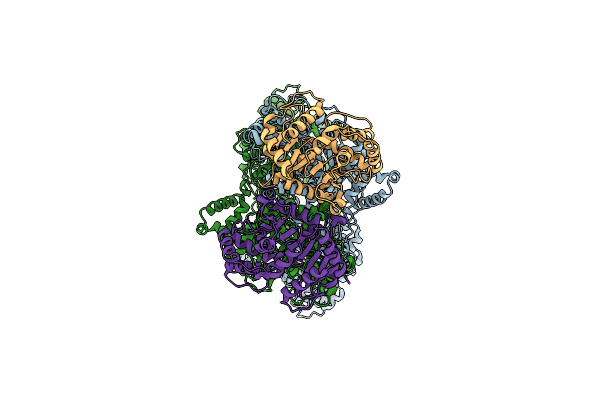
Crystal Structure Of Nanobody Tnb04-1 With Antibody 1F11 Fab And Sars-Cov-2 Rbd
Organism: Vicugna pacos, Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
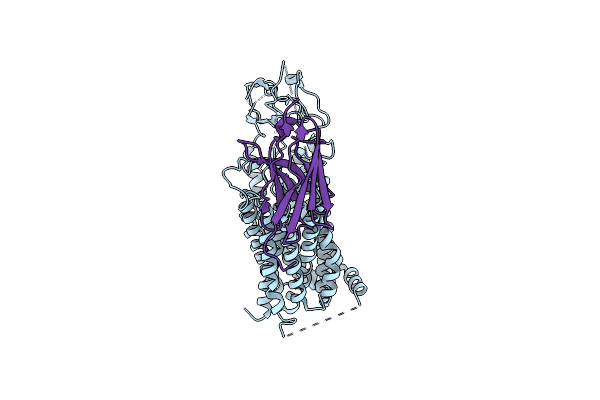 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: TRANSPORT PROTEIN |
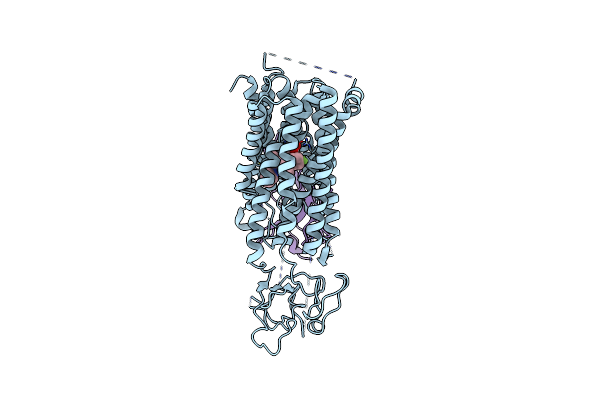 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: TRANSPORT PROTEIN Ligands: 117 |
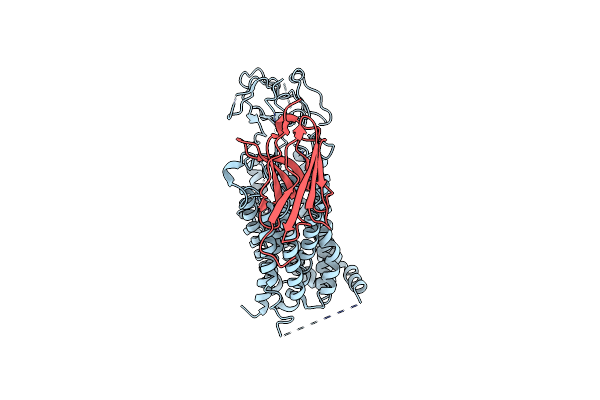 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: TRANSPORT PROTEIN |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |

