Search Count: 1,368
 |
Structures And Mechanism Of The Nicotinamide Nucleotide Transhydrogenase From E. Coli
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: MEMBRANE PROTEIN Ligands: PC1, PKZ |
 |
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, A1BWO, ZN |
 |
Crystal Structure Of Hla-A*11:01 In Complex With Kras G12D 9-Mer Peptide (Vvgadgvgk)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: IMMUNE SYSTEM |
 |
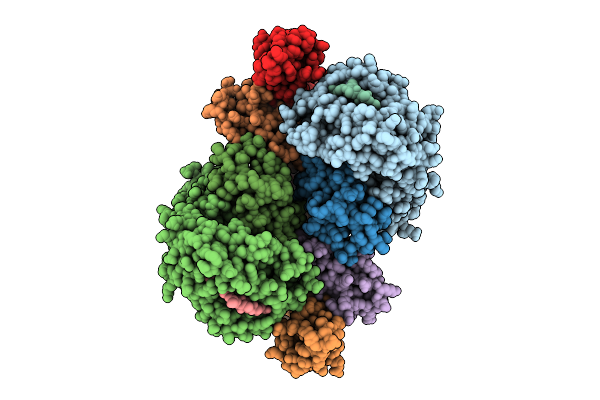
Crystal Structure Of Hla-A*11:01 In Complex With Kras G12A 10-Mer Peptide (Vvvgaagvgk)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: IMMUNE SYSTEM |
 |
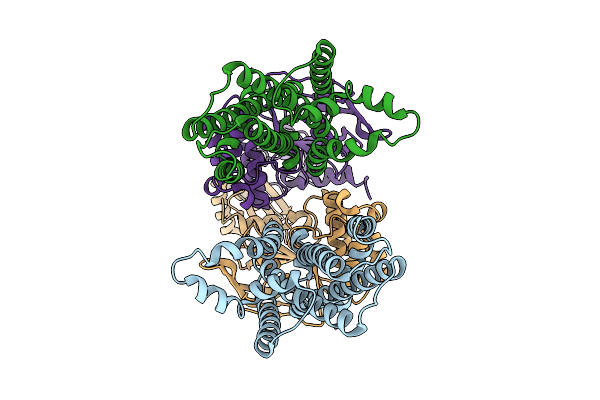
Crystal Structure Of Hla-A*11:01 In Complex With Kras G12S 10-Mer Peptide (Vvvgasgvgk)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: IMMUNE SYSTEM |
 |
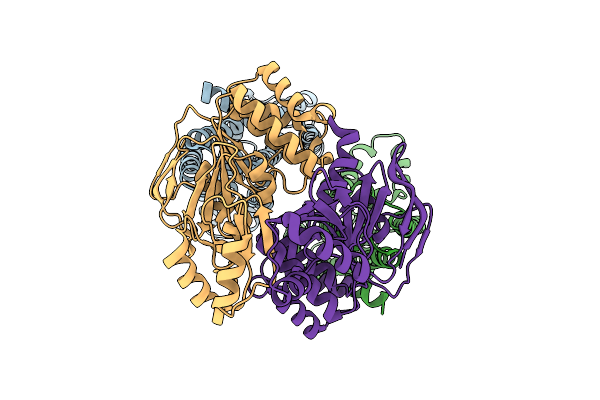
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
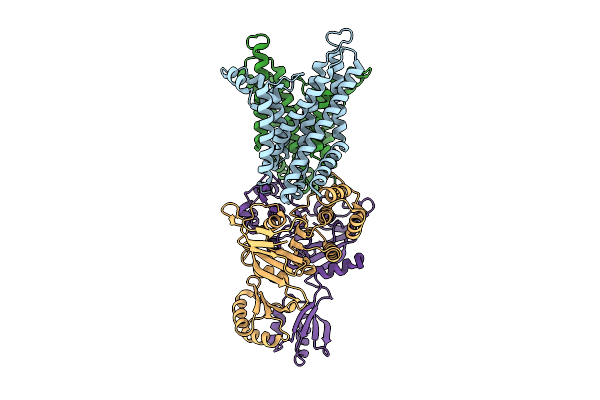 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
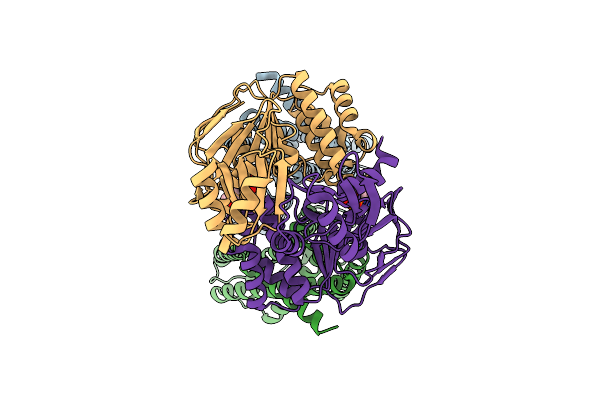 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: ADP |
 |
Cryo-Em Structure Of Ternary Complex Bcl6-Crbn-Ddb1 With Bcl6-760 (Ldd, Local Refined)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: LIGASE Ligands: ZN, A1CM7 |
 |
Organism: Entacmaea quadricolor
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: FLUORESCENT PROTEIN Ligands: EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LIGASE Ligands: A1CNP, ZN |
 |
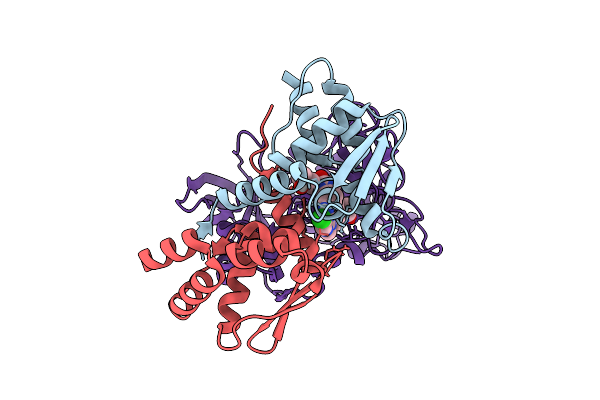
Cryo-Em Structure Of Ternary Complex Ikaros-Zf2:Cc-885:Crbn:Ddb1 (Molecular Glue Degrader)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: LIGASE Ligands: ZN, 85C |
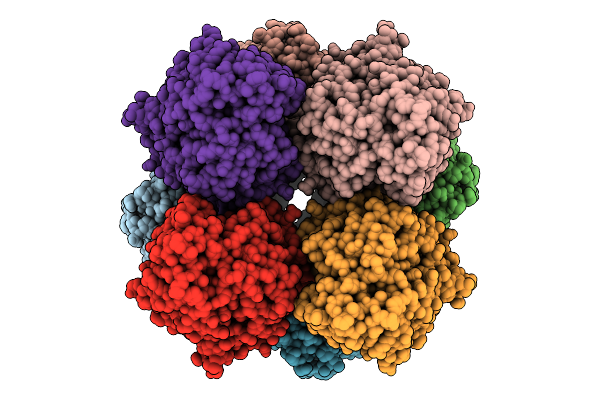 |
Structure Of Lara-Like Nickel-Pincer Nucleotide Cofactor-Utilizing Enzyme With A Single Catalytic Histidine Residue From Streptococcus Plurextorum
Organism: Streptococcus plurextorum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: ISOMERASE Ligands: NI, 4EY |
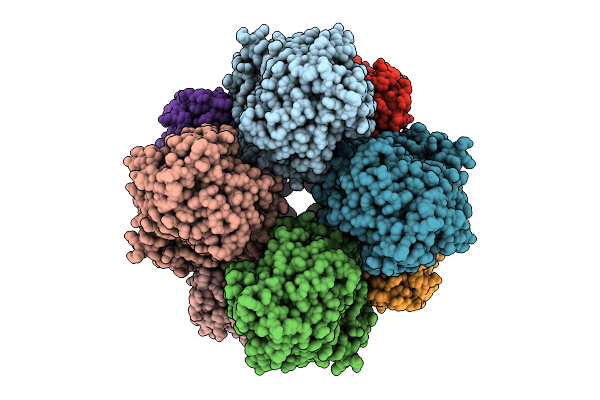 |
Structures Of Lara-Like Nickel-Pincer Nucleotide Cofactor-Utilizing Enzyme With A Single Catalytic Histidine Residue From Blautia Wexlerae
Organism: Blautia wexlerae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: ISOMERASE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: HYDROLASE Ligands: GTP, MG, NA |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: HYDROLASE Ligands: GTP, MG, K |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: HYDROLASE Ligands: GDP, BEF, MG, K, MES |
 |
Structure Of An Ancestral Ethylene Forming Enzyme, Anc357, In Complex With Mn
Organism: Unidentified
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: OXIDOREDUCTASE Ligands: MN |

