Search Count: 519
 |
Organism: Psychrobacter lutiphocae dsm 21542, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2026-02-04 Classification: RNA BINDING PROTEIN/RNA Ligands: CA, GOL, PEG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: LIGASE Ligands: ZN, QFC |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: LIGASE Ligands: ZN, QFC |
 |
Organism: Psychrobacter lutiphocae dsm 21542, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2026-02-04 Classification: RNA BINDING PROTEIN/RNA Ligands: MG |
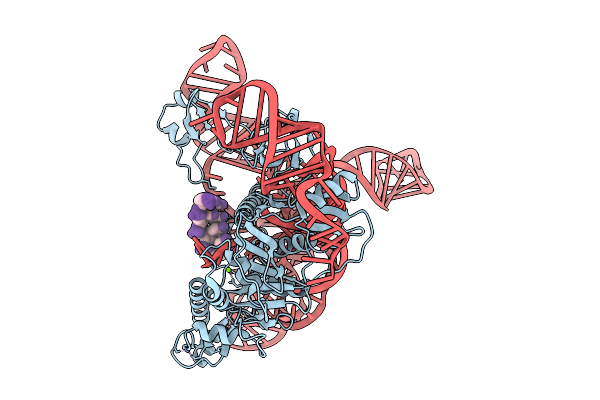 |
Cryoem Structures Uncover The Unexpected Hinges Of Iscb For Enhanced Gene Editing
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.04 Å Release Date: 2026-01-21 Classification: RNA BINDING PROTEIN Ligands: ZN, MG |
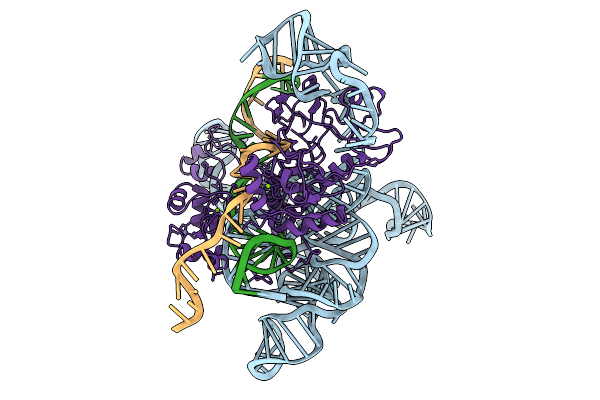 |
Cryoem Structures Uncover The Unexpected Hinges Of Iscb For Enhanced Gene Editing
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2026-01-21 Classification: DNA BINDING PROTEIN Ligands: MG, ZN |
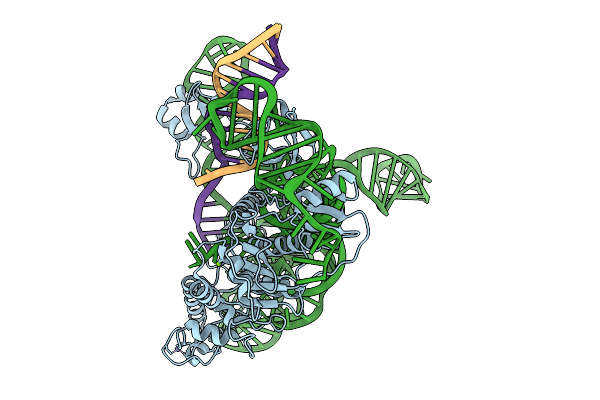 |
Cryoem Structures Uncover The Unexpected Hinges Of Iscb For Enhanced Gene Editing
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.39 Å Release Date: 2026-01-21 Classification: RNA BINDING PROTEIN Ligands: ZN, MG |
 |
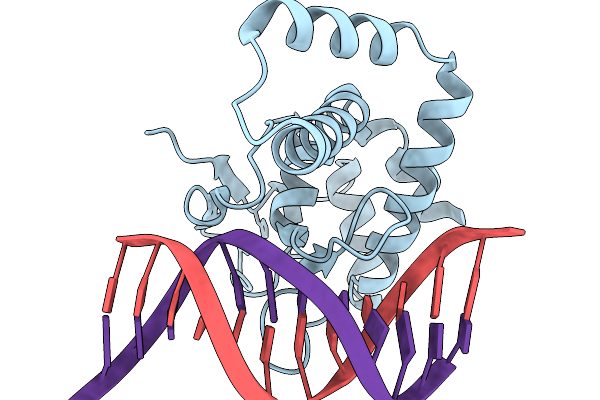
Cryoem Structures Uncover The Unexpected Hinges Of Iscb For Enhanced Gene Editing
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.09 Å Release Date: 2026-01-21 Classification: DNA BINDING PROTEIN Ligands: ZN, MG |
 |
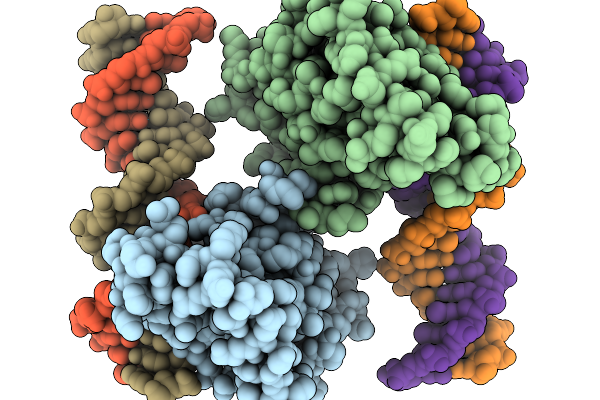
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2026-01-07 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
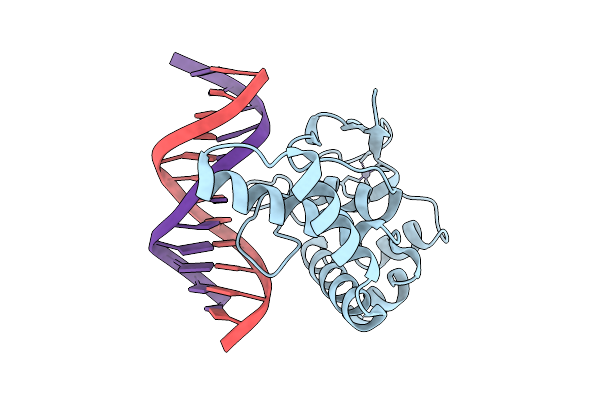
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2026-01-07 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
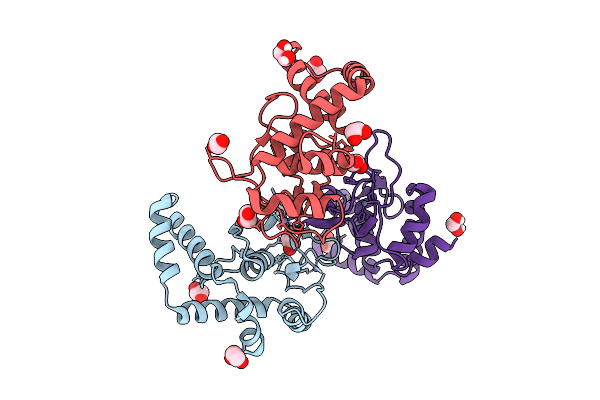
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
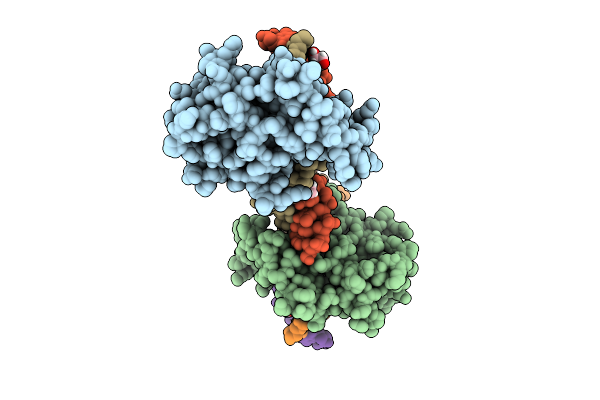
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN, EDO |
 |
Crystal Structure Of Nfia In Complex With Dna Containing The Tggca(N3)Tgcca Motif
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN, EDO |
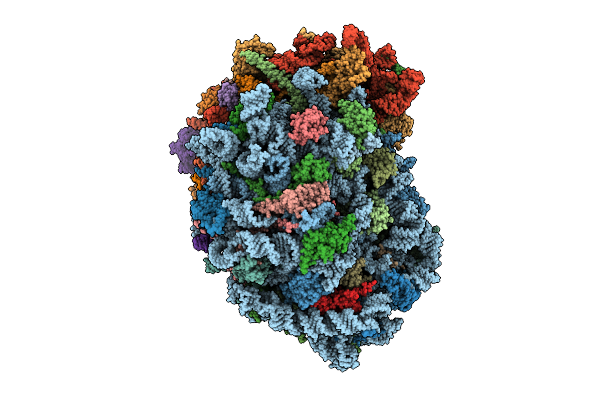 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
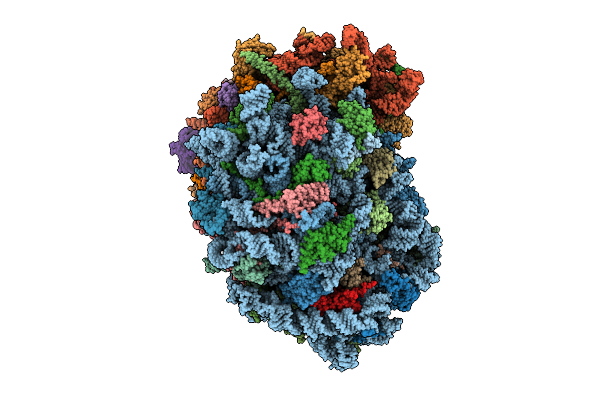 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
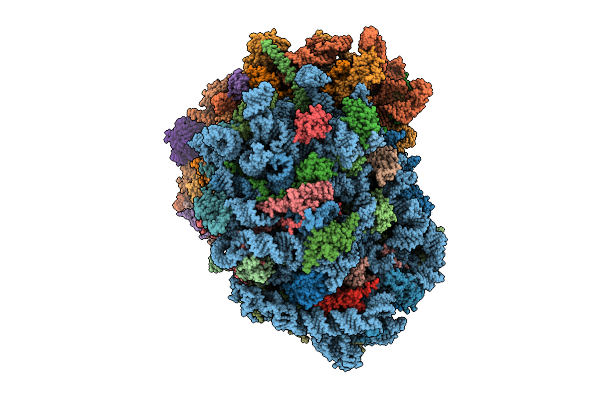 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, ZN, SPM, SPD |
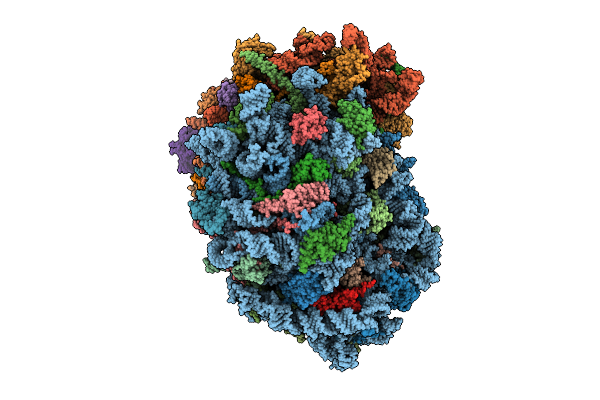 |
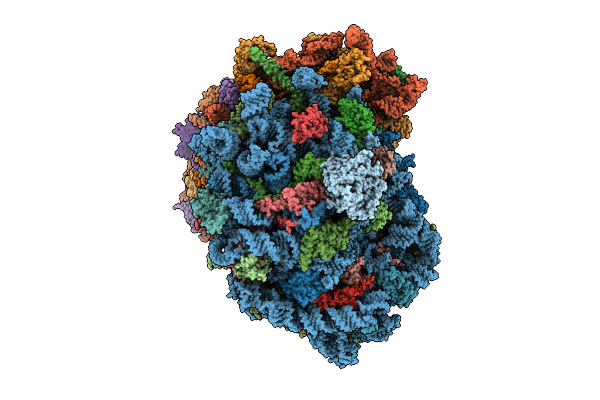
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
 |
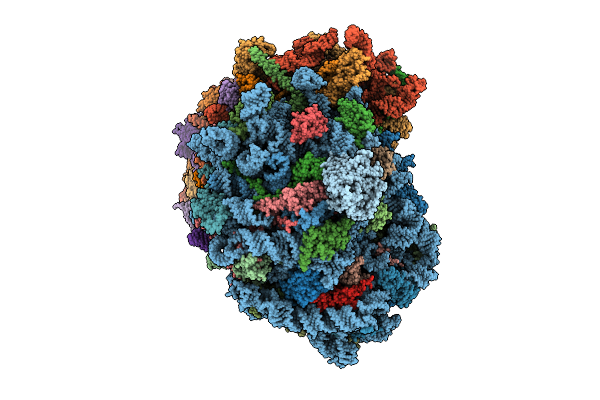
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
 |
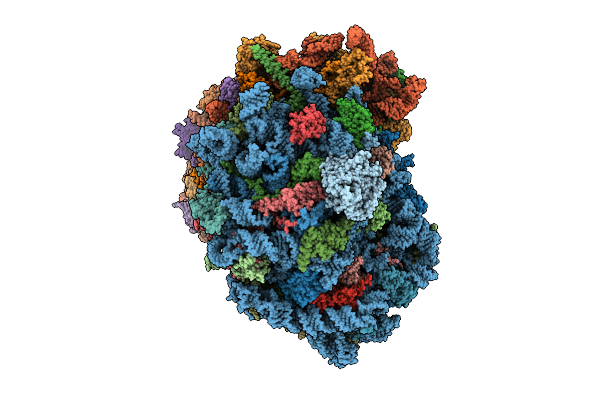
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |

