Search Count: 265
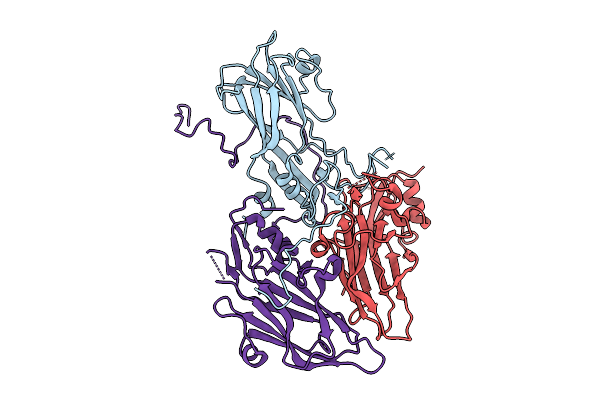 |
Organism: Coxsackievirus a6
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRUS |
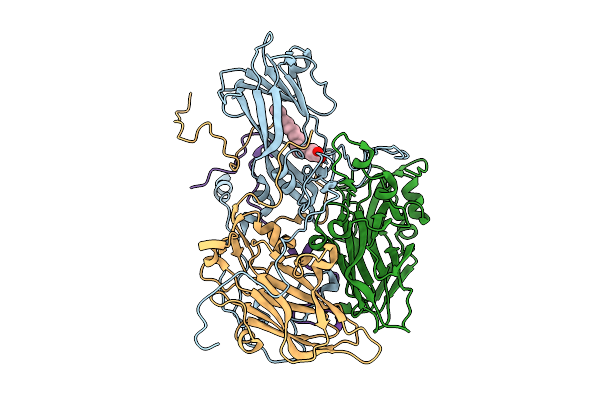 |
Organism: Coxsackievirus a6
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRUS Ligands: STE |
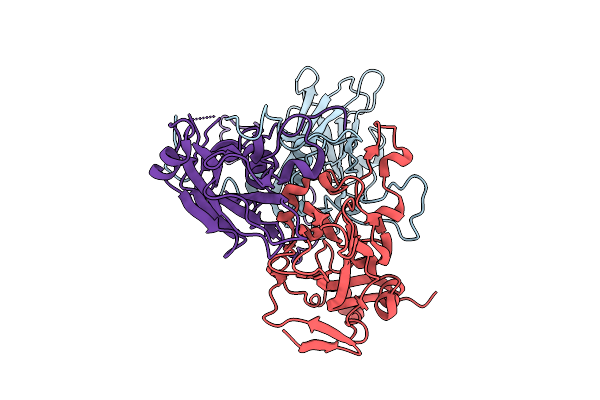 |
Organism: Coxsackievirus a6
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRUS |
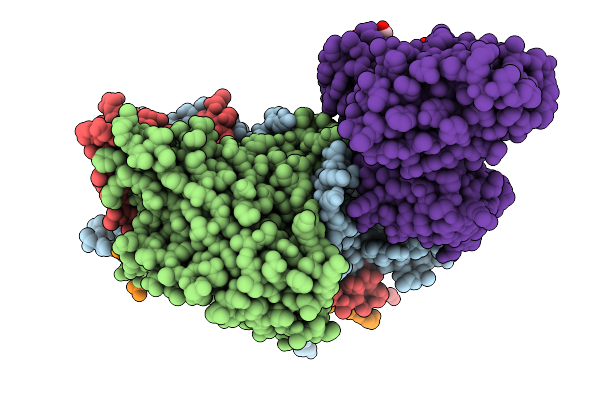 |
Structure Of Mature Coxsackievirus A6 Virion Complexed With Its Receptor Kremen1
Organism: Homo sapiens, Coxsackievirus a6
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: VIRUS Ligands: STE, MYR, NAG |
 |
Organism: Mus musculus, Coxsackievirus a6
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: VIRUS Ligands: STE |
 |
Organism: Mus musculus, Coxsackievirus a6
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: VIRUS Ligands: STE |
 |
Organism: Mus musculus, Coxsackievirus a6
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: VIRUS |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PT5, K, CA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PIO, K |
 |
Organism: Homo sapiens, Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: GENE REGULATION Ligands: ZN |
 |
Structure Of Human Line-1 Orf2P With Endogenous Dna And Rna/Cdna Hybrid Bound To Dntp And Mn2+
Organism: Homo sapiens, Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: GENE REGULATION Ligands: DTP, ZN, MN |
 |
Structure Of Full-Length Human Line-1 Orf2P With Endogenous Dna And Rna/Cdna Hybrid
Organism: Homo sapiens, Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: GENE REGULATION Ligands: ZN |
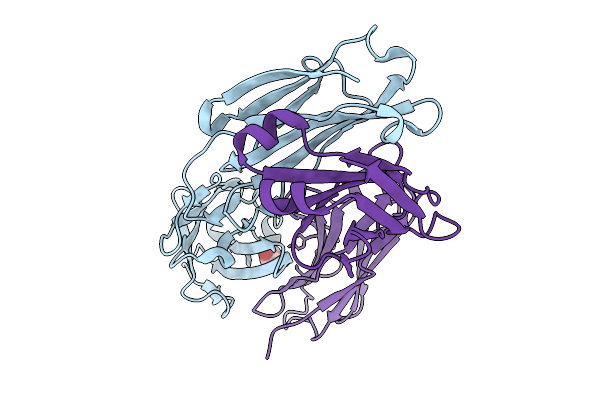 |
Crystal Structure Of Switchbody Based On Anti-Osteocalcin Antibody Ktm219 Fab
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: IMMUNE SYSTEM Ligands: GOL |
 |
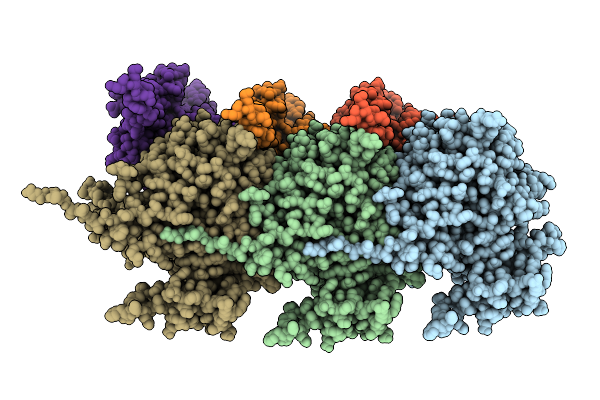
Organism: Zika virus zikv/h. sapiens/frenchpolynesia/10087pf/2013, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: VIRUS/IMMUNE SYSTEM |
 |
Organism: Porphyromonas gingivalis atcc 33277
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: VIRAL PROTEIN |
 |
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: HYDROLASE Ligands: A1EBP, GOL, BEZ, PDO, BU1, PGO |
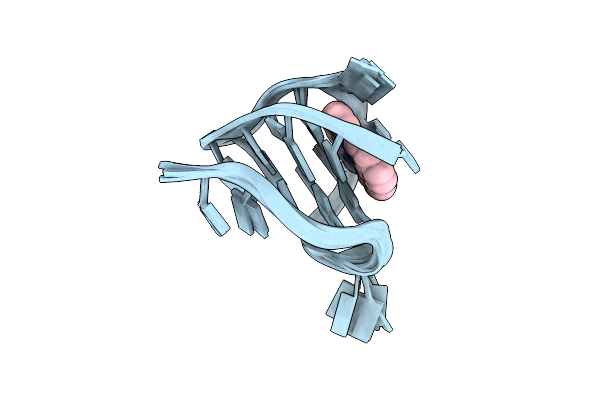 |
Nmr Solution Structure Of The 1:1 Complex Of A Platinum(Ii) Compound Bound To Myc1234 G-Quadruplex
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-05-07 Classification: DNA Ligands: H9C |
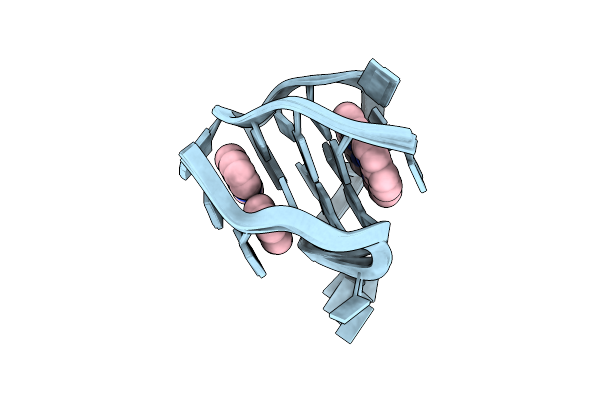 |
Nmr Solution Structure Of The 2:1 Complex Of A Platinum(Ii) Compound Bound To Myc1234 G-Quadruplex
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-04-09 Classification: DNA Ligands: H9C |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2025-03-19 Classification: VIRAL PROTEIN/RNA Ligands: ZN, MG |

