Search Count: 32
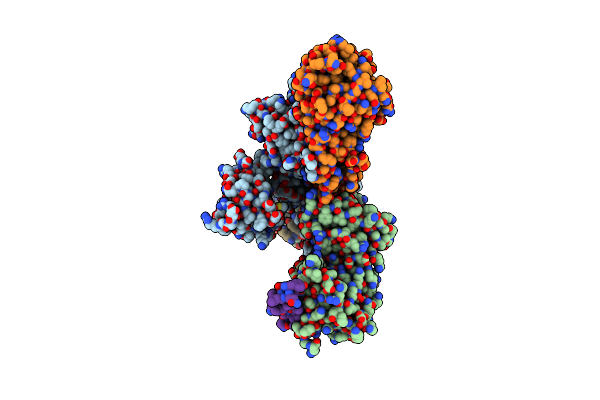 |
Structure Of The C-Terminal Protease Ctpa-Lbca Complex Of Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: HYDROLASE |
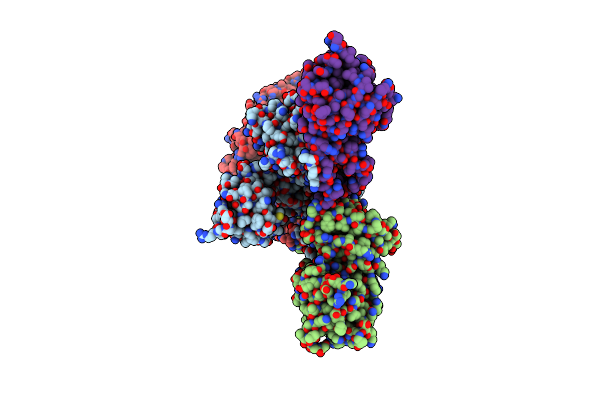 |
The C-Terminal Protease Ctpa-Lbca Complex Of Pseudomonas Aeruginosa With The Tpr At The High Position
Organism: Pseudomonas aeruginosa, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: HYDROLASE |
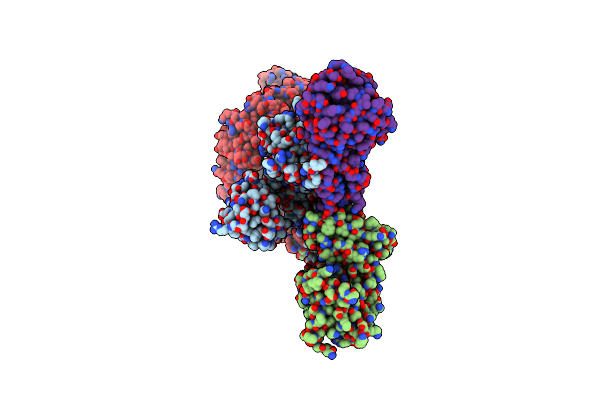 |
The C-Terminal Protease Ctpa-Lbca Complex Of Pseudomonas Aeruginosa With The Tpr At The Low Position
Organism: Pseudomonas aeruginosa, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: HYDROLASE |
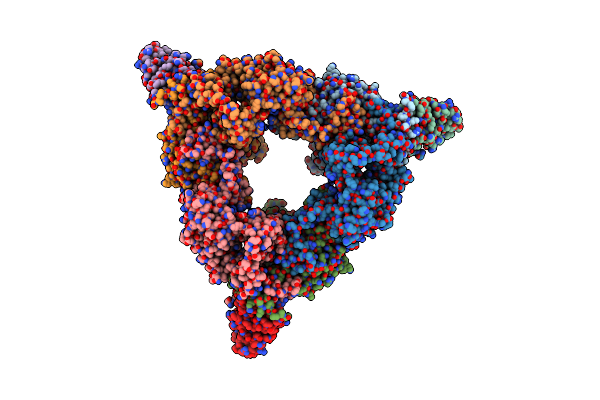 |
Structure Of The C-Terminal Protease Ctpa-Lbca Complex Of Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: HYDROLASE |
 |
Structure Of The Plasmodium Falciparum 20S Proteasome Complexed With Inhibitor Tdi-8304
Organism: Plasmodium falciparum nf54
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: YRE |
 |
Structure Of The Plasmodium Falciparum 20S Proteasome Beta-6 A117D Mutant Complexed With Inhibitor Wlw-Vs
Organism: Plasmodium falciparum dd2
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 7F1 |
 |
Structure Of Human Constitutive 20S Proteasome Complexed With The Inhibitor Tdi-8304
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: YRE |
 |
 |
 |
Crystal Structure Of Carboxyl-Terminal Processing Protease A, Ctpa, Of Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2022-04-27 Classification: HYDROLASE |
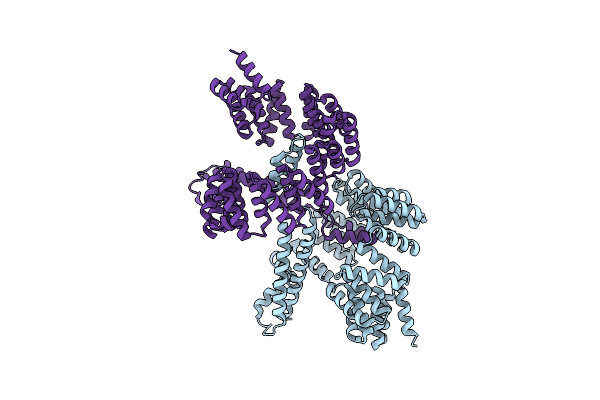 |
Crystal Structure Of Lbca (Lipoprotein Binding Partner Of Ctpa) Of Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2022-04-27 Classification: PROTEIN BINDING |
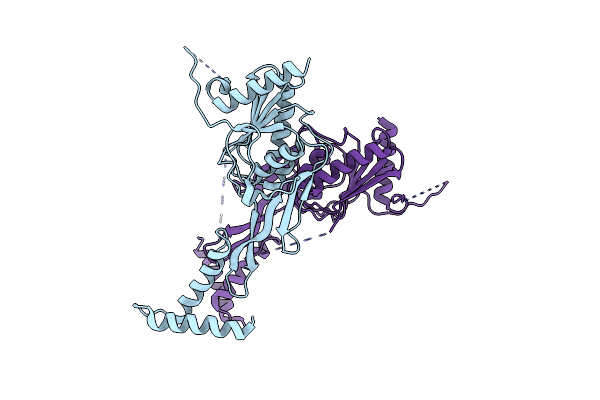 |
Crystal Structure Of Carboxyl-Terminal Processing Protease A Mutant S302A, Ctpa_S302A, Of Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-04-27 Classification: HYDROLASE |
 |
Macrocyclic Peptides Tdi5575 That Selectively Inhibit The Mycobacterium Tuberculosis Proteasome
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2021-04-28 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: U5Y, CIT, DMF |
 |
Crystal Structure Of Mycobacterium Tuberculosis Proteasome In Complex With Phenylimidazole-Based Inhibitor A85
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-10-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: DMF, M6M, CIT |
 |
Crystal Structure Of Mycobacterium Tuberculosis Proteasome In Complex With Phenylimidazole-Based Inhibitor A86
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2019-10-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: DMF, M6Y, CIT |
 |
Crystal Structure Of Mycobacterium Tuberculosis Proteasome In Complex With Phenylimidazole-Based Inhibitor B6
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-10-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: M9G |
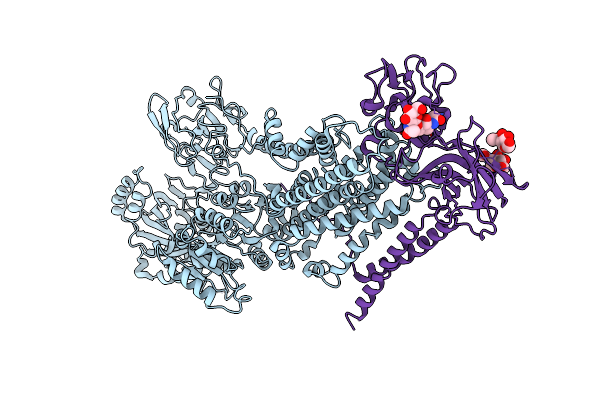 |
Organism: Saccharomyces cerevisiae w303
Method: ELECTRON MICROSCOPY Release Date: 2019-09-25 Classification: TRANSLOCASE Ligands: NAG |
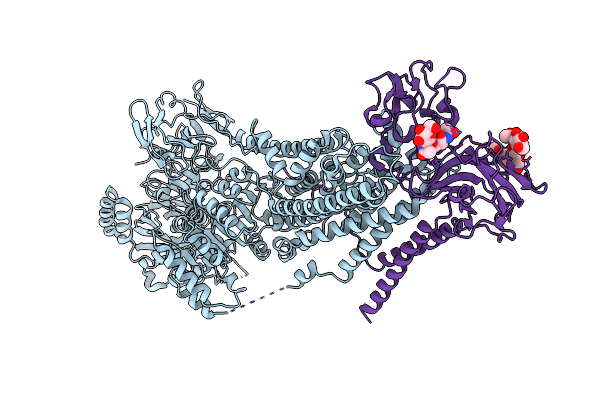 |
Cryo-Em Structure Of S. Cerevisiae Drs2P-Cdc50P In The Autoinhibited Apo Form
Organism: Saccharomyces cerevisiae w303
Method: ELECTRON MICROSCOPY Release Date: 2019-09-25 Classification: TRANSLOCASE Ligands: NAG |
 |
Crystal Structure Of Tetr Family Regulator Rv0078 From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-07-04 Classification: TRANSCRIPTION Ligands: SO4 |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv), Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-07-04 Classification: DNA BINDING PROTEIN/DNA |

