Search Count: 67
 |
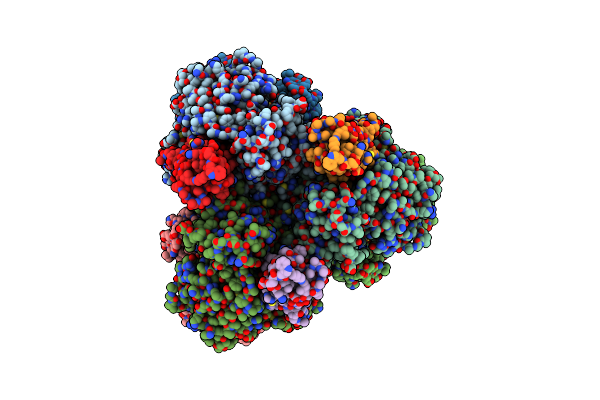
Cryo-Em Structure Of Cryptococcus Neoformans Trehalose-6-Phosphate Synthase Homotetramer In Complex With Uridine Diphosphate And Glucose-6-Phosphate
Organism: Cryptococcus neoformans var. grubii h99
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: TRANSFERASE Ligands: UDP, G6P |
 |
Organism: Escherichia coli str. k-12 substr. mg1655, Escherichia phage t7
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: HYDROLASE |
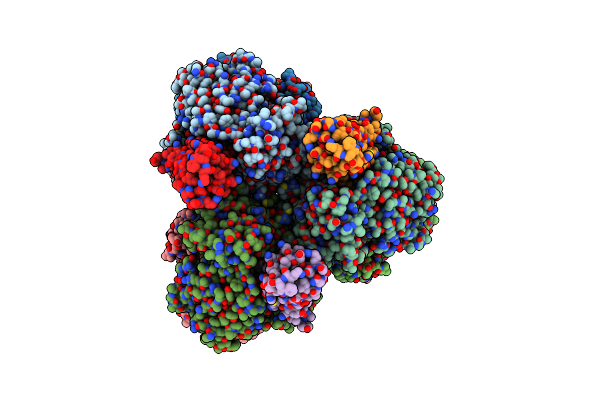 |
Structure Of E. Coli Dgtpase Bound To T7 Bacteriophage Protein Gp1.2 And Dgtp
Organism: Escherichia coli str. k-12 substr. mg1655, Escherichia phage t7
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: HYDROLASE Ligands: MG, DGT |
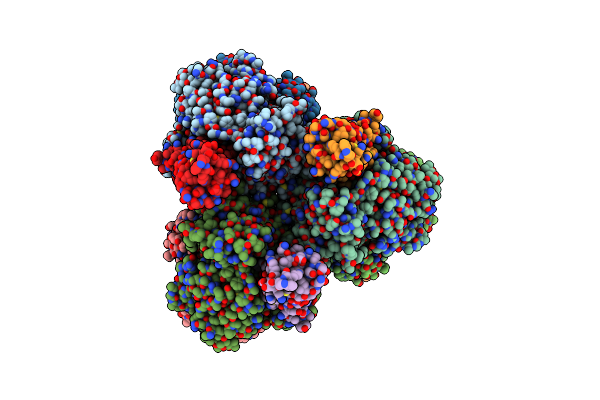 |
Structure Of E. Coli Dgtpase Bound To T7 Bacteriophage Protein Gp1.2 And Gtp
Organism: Escherichia coli str. k-12 substr. mg1655, Escherichia phage t7
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: HYDROLASE Ligands: GTP, MG |
 |
Organism: Leeuwenhoekiella blandensis
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2022-06-01 Classification: HYDROLASE Ligands: SO4, MN |
 |
Organism: Leeuwenhoekiella blandensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-06-01 Classification: HYDROLASE Ligands: EDO, SO4 |
 |
Organism: Leeuwenhoekiella blandensis
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2022-06-01 Classification: HYDROLASE Ligands: SO4, MN |
 |
Organism: Leeuwenhoekiella blandensis
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2022-06-01 Classification: HYDROLASE Ligands: EDO, SO4, MG |
 |
Organism: Leeuwenhoekiella blandensis
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2022-06-01 Classification: HYDROLASE Ligands: EDO, SO4, MN |
 |
Organism: Leeuwenhoekiella blandensis med217
Method: ELECTRON MICROSCOPY Release Date: 2022-06-01 Classification: HYDROLASE Ligands: MG |
 |
Organism: Leeuwenhoekiella blandensis med217
Method: ELECTRON MICROSCOPY Release Date: 2022-06-01 Classification: HYDROLASE Ligands: DTP, MG |
 |
Organism: Leeuwenhoekiella blandensis med217
Method: ELECTRON MICROSCOPY Release Date: 2022-06-01 Classification: HYDROLASE Ligands: DGT, MG |
 |
Organism: Leeuwenhoekiella blandensis med217
Method: ELECTRON MICROSCOPY Release Date: 2022-06-01 Classification: HYDROLASE Ligands: DGT, DTP, MG |
 |
Organism: Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2021-04-14 Classification: VIRAL PROTEIN Ligands: NAG |
 |
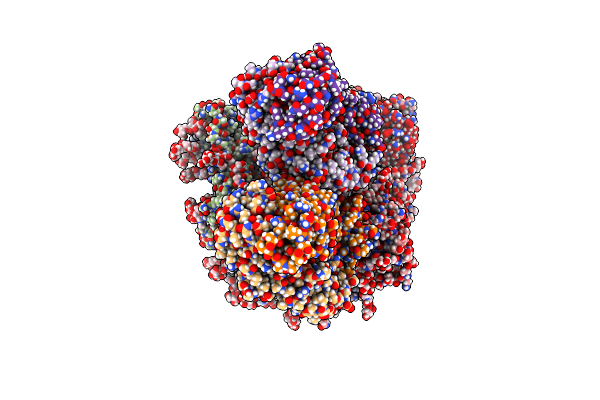
Organism: Macaca mulatta, Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2021-03-24 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Dh898.1 Fab-Dimer Bound Near The Cd4 Binding Site Of Hiv-1 Env Ch848 Sosip Trimer
Organism: Human immunodeficiency virus 1, Simian-human immunodeficiency virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-03-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Dh898.1 Fab-Dimer From Local Refinement Of The Fab-Dimer Bound Near The Cd4 Binding Site Of Hiv-1 Env Ch848 Sosip Trimer
Organism: Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2021-02-10 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2020-12-30 Classification: IMMUNE SYSTEM Ligands: GOL |
 |
Crystal Structure Of The Disulfide Linked Dh717.1 Fab Dimer, Derived From A Macaque Hiv-1 Vaccine-Induced Env Glycan-Reactive Neutralizing Antibody B Cell Lineage
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2020-12-30 Classification: IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Sars-Cov-2 2P S Ectodomain Bound To One Copy Of Domain-Swapped Antibody 2G12
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-30 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |

