Search Count: 59
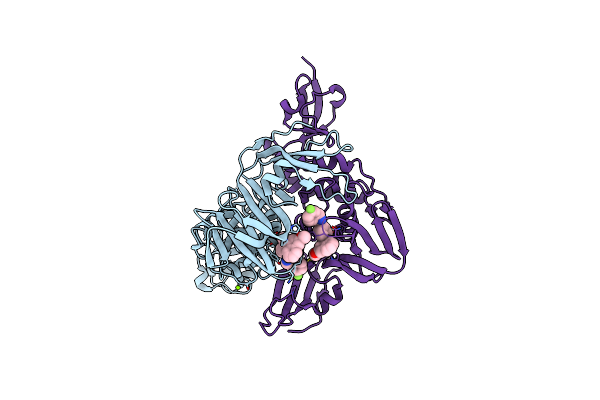 |
Crystal Structure Of Sars-Cov-2 Papain-Like Protease Complexed With Noncovalent Inhibitor Sr-01
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-07-03 Classification: VIRAL PROTEIN/INHIBITOR Ligands: MG, V00 |
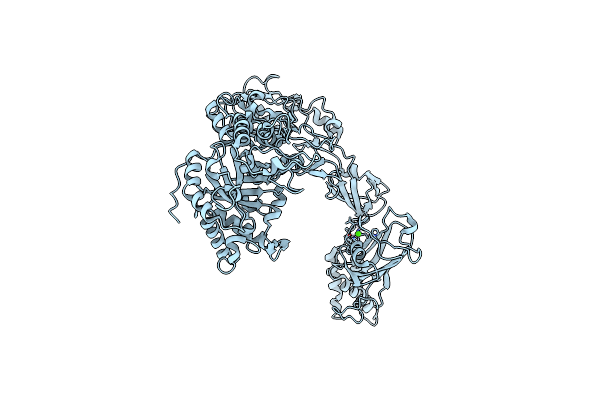 |
Crystal Structure Of Endosz Mutant D234M, From Streptococcus Equi Subsp. Zooepidemicus Sz105
Organism: Streptococcus equi subsp. zooepidemicus sz105
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-07-03 Classification: HYDROLASE Ligands: CA |
 |
Organism: Streptococcus equi subsp. zooepidemicus sz105
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-07-03 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Closed Conformation Of Human Immunoglobulin Fc In Presence Of Endosz
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2024-07-03 Classification: IMMUNE SYSTEM Ligands: CL |
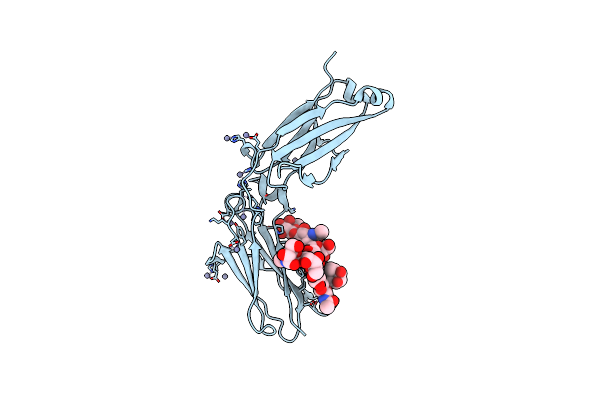 |
Crystal Structure Of Open Conformation Of Human Immunoglobulin Fc In Presence Of Endosz
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2024-07-03 Classification: IMMUNE SYSTEM Ligands: ZN |
 |
Crystal Structure Of Endosz Mutant D234M, In Space Group P21, In Complex With Oligosaccharide G2S1
Organism: Streptococcus equi subsp. zooepidemicus sz105
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2024-07-03 Classification: HYDROLASE Ligands: CA |
 |
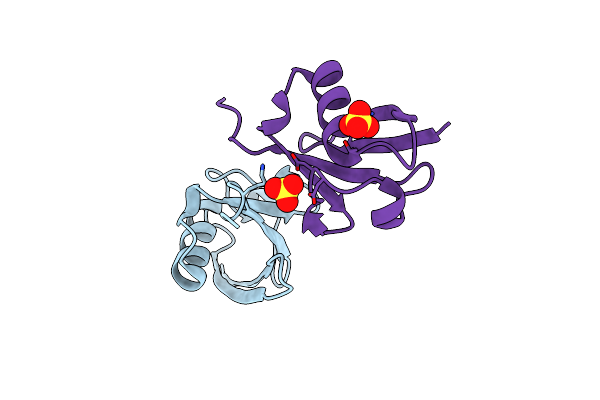
Crystal Structure Of Endosz Mutant D234M, From Streptococcus Equi Subsp. Zooepidemicus Sz105, In Complex With Oligosaccharide G2S2-Oxazoline
Organism: Streptococcus equi subsp. zooepidemicus sz105
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2024-07-03 Classification: HYDROLASE Ligands: NGO, CA |
 |
Robust Design Of Effective Allosteric Activator Ubv R4 For Rsp5 E3 Ligase Using The Machine-Learning Tool Proteinmpnn
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-08-09 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4 |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:4.20 Å Release Date: 2023-02-01 Classification: METAL BINDING PROTEIN |
 |
Crystal Structure Of Pwwp-Dna Complex For Human Hepatoma-Derived Growth Factor
Organism: Escherichia coli bl21, Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2018-06-20 Classification: HORMONE Ligands: MPD |
 |
Crystal Structure Of The Pwwp Domain Of Human Hepatoma-Derived Growth Factor
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2018-06-20 Classification: HORMONE |
 |
Organism: Desulfovibrio gigas, Desulfovibrio gigas dsm 1382 = atcc 19364
Method: X-RAY DIFFRACTION Release Date: 2018-06-06 Classification: ELECTRON TRANSPORT Ligands: FAD, FUM, F3S, SF4, FES, HEM, LMT, MQ7 |
 |
The N253F Mutant Structure Of Trehalose Synthase From Deinococcus Radiodurans Reveals An Open Active-Site Conformation
Organism: Deinococcus radiodurans str. r1
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2017-10-25 Classification: ISOMERASE Ligands: CA, MG |
 |
Truncated Human Tifa In Complex With Its Thr9 Phosphorylated N-Terminal Peptide 1-15
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2015-10-21 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-10-14 Classification: SIGNALING PROTEIN |
 |
Crystal Structures Of Trehalose Synthase From Deinococcus Radiodurans Reveal That A Closed Conformation Is Involved In The Intramolecular Isomerization Catalysis
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2014-12-24 Classification: ISOMERASE Ligands: CA, MG, TRS |
 |
Crystal Structure Of Trehalose Synthase From Deinococcus Radiodurans Reveals A Closed Conformation For Catalysis Of The Intramolecular Isomerization
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-12-17 Classification: ISOMERASE Ligands: CA, MG, TRS |
 |
Organism: Tetraodon nigroviridis
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2013-09-18 Classification: HYDROLASE ACTIVATOR Ligands: ZN, URQ |
 |
Organism: Tetraodon nigroviridis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-09-18 Classification: HYDROLASE ACTIVATOR Ligands: URP, ZN |
 |
The Crystal Structure Of Di-Zn Dihydropyrimidinase In Complex With Hydantoin
Organism: Tetraodon nigroviridis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-09-18 Classification: HYDROLASE ACTIVATOR Ligands: ZN, HYN |

