Search Count: 31
 |
Catalytic And Non-Catalytic Mechanisms Of Histone H4 Lysine 20 Methyltransferase Suv420H1
Organism: Xenopus laevis, Homo sapiens, Escherichia coli 'bl21-gold(de3)plyss ag'
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: GENE REGULATION |
 |
Catalytic And Non-Catalytic Mechanisms Of Histone H4 Lysine 20 Methyltransferase Suv420H1
Organism: Homo sapiens, Xenopus laevis, Escherichia coli 'bl21-gold(de3)plyss ag'
Method: ELECTRON MICROSCOPY Release Date: 2023-09-06 Classification: GENE REGULATION Ligands: SAM |
 |
Catalytic And Non-Catalytic Mechanisms Of Histone H4 Lysine 20 Methyltransferase Suv420H1
Organism: Homo sapiens, Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-09-06 Classification: GENE REGULATION/DNA |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-07-19 Classification: DNA BINDING PROTEIN Ligands: ADP |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-07-19 Classification: DNA BINDING PROTEIN Ligands: ADP |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-07-19 Classification: DNA BINDING PROTEIN Ligands: ADP |
 |
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: STRUCTURAL PROTEIN/DNA |
 |
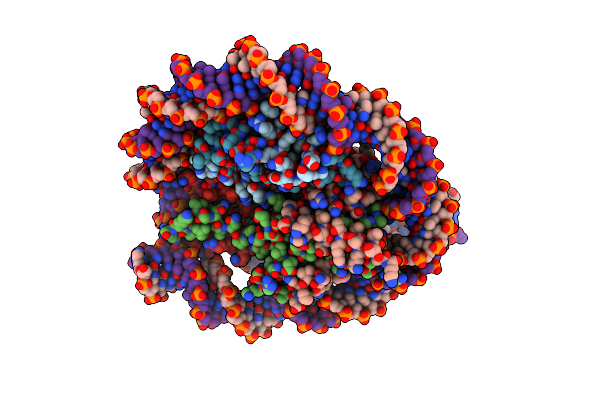
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.16 Å Release Date: 2023-07-12 Classification: MOTOR PROTEIN/DNA |
 |
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: STRUCTURAL PROTEIN/DNA |
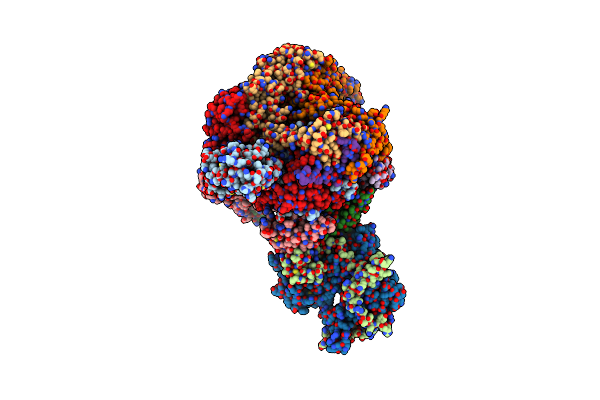 |
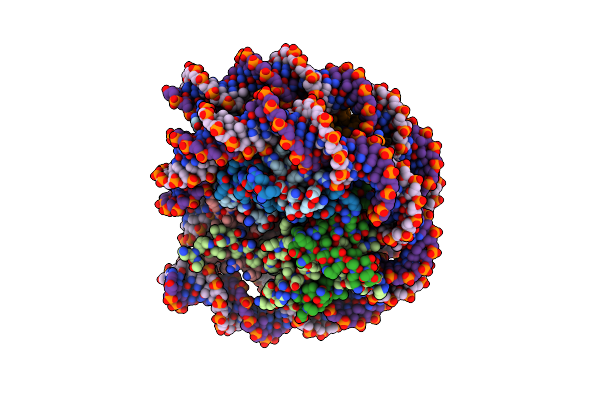
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: DNA BINDING PROTEIN/Hydrolase Ligands: ADP |
 |
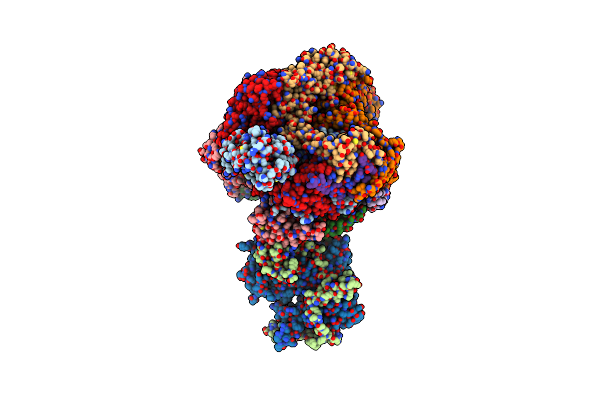
Organism: Xenopus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: DNA BINDING PROTEIN/Hydrolase Ligands: ADP |
 |
Organism: Xenopus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2018-10-17 Classification: ANTIBIOTICS/DNA Ligands: CPH, CO |
 |
Crystal Structure Of The Murine Norovirus Vp1 P Domain In Complex With The Cd300Lf Receptor
Organism: Murine norovirus 1, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-09-12 Classification: VIRAL PROTEIN/LIPID BINDING PROTEIN Ligands: EDO, CL, MG |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2018-09-12 Classification: LIPID BINDING PROTEIN Ligands: PC, CA |
 |
Crystal Structure Of The Murine Norovirus Vp1 P Domain In Complex With The Cd300Lf Receptor And Glycochenodeoxycholic Acid
Organism: Murine norovirus 1, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-09-12 Classification: VIRAL PROTEIN Ligands: MG, EDO, CHO |
 |
Crystal Structure Of The Murine Norovirus Vp1 P Domain In Complex With The Cd300Lf Receptor And Lithocholic Acid
Organism: Murine norovirus 1, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-09-12 Classification: VIRAL PROTEIN Ligands: 4OA, CA, EDO |
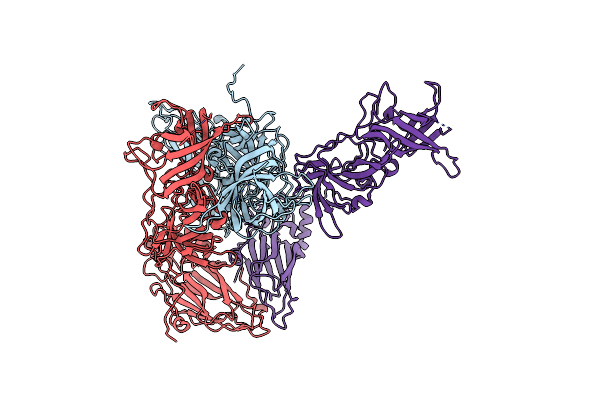 |
Mouse Norovirus Model Using The Crystal Structure Of Mnv P Domain And The Norwalkvirus Shell Domain
Organism: Norwalk virus, Murine norovirus 1
Method: ELECTRON MICROSCOPY Release Date: 2018-04-04 Classification: VIRUS |
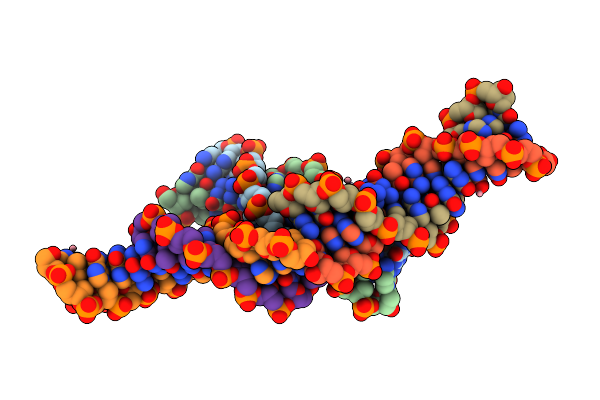 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2017-08-30 Classification: DNA Ligands: CO |

