Search Count: 19
 |
Organism: Thermobifida cellulosilytica
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-07-12 Classification: HYDROLASE Ligands: PEG, MG, CL |
 |
Organism: Thermobifida cellulosilytica
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-07-12 Classification: HYDROLASE Ligands: CL |
 |
Structure Of A Double Variant Of Cutinase 2 From Thermobifida Cellulosilytica
Organism: Thermobifida cellulosilytica
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2017-07-12 Classification: HYDROLASE Ligands: CL, MG |
 |
Structure Of A Triple Variant Of Cutinase 2 From Thermobifida Cellulosilytica
Organism: Thermobifida cellulosilytica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-07-12 Classification: HYDROLASE Ligands: CA, CL |
 |
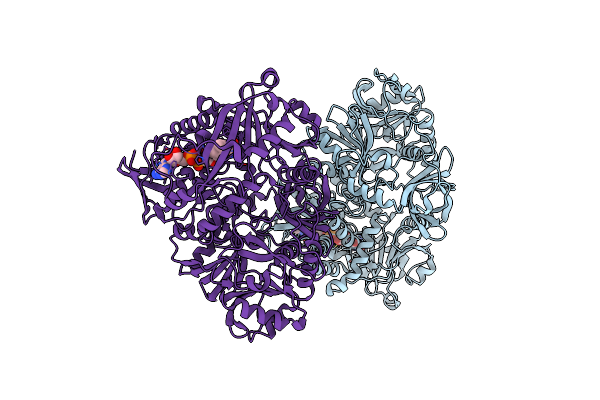
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2017-05-24 Classification: OXIDOREDUCTASE Ligands: PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2016-08-17 Classification: OXIDOREDUCTASE Ligands: FAD |
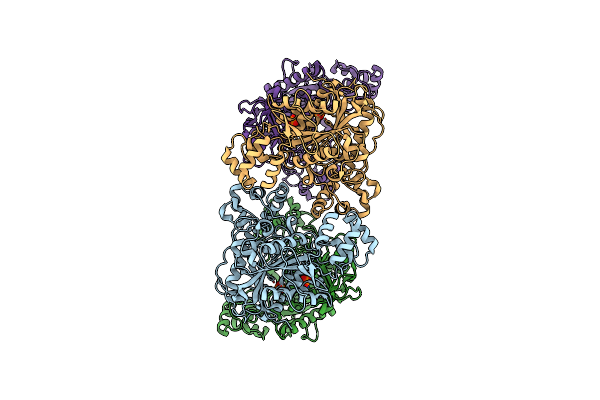 |
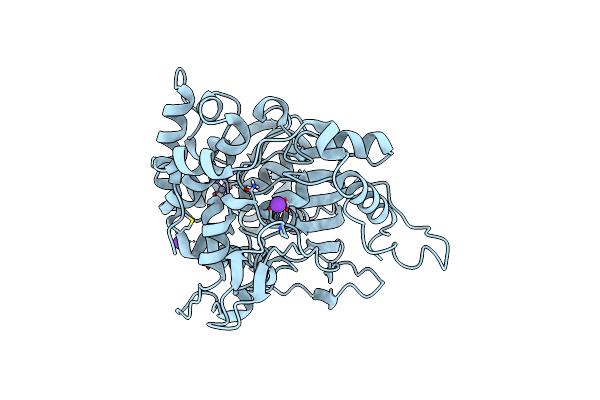
An Esterase From Anaerobic Clostridium Hathewayi Can Hydrolyze Aliphatic Aromatic Polyesters
Organism: Hungatella hathewayi
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-02-24 Classification: HYDROLASE Ligands: PO4 |
 |
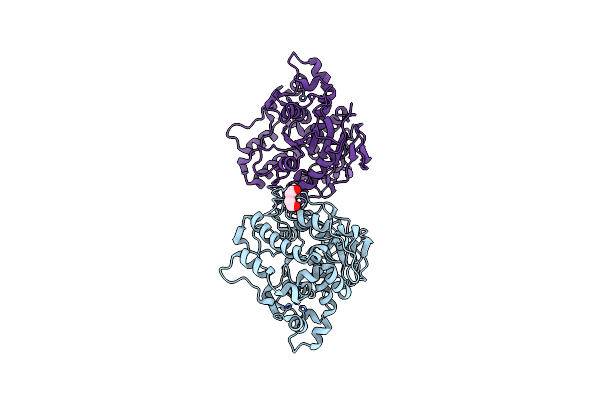
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2015-11-11 Classification: HYDROLASE Ligands: ZN, K |
 |
Organism: Pelosinus fermentans dsm 17108
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-11-04 Classification: HYDROLASE Ligands: ZN, PEG, K |
 |
Organism: Elizabethkingia meningoseptica
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2015-07-01 Classification: LYASE Ligands: FAD, NA, P6G, PO4 |
 |
Organism: Enterobacter sp.
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2015-06-10 Classification: LYASE Ligands: PO4, GLY, SO4 |
 |
Organism: Enterobacter sp.
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2015-06-10 Classification: LYASE |
 |
Organism: Eschscholzia californica
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-04-23 Classification: OXIDOREDUCTASE Ligands: FAD, NAG, SO4, 12P |
 |
Organism: Phleum pratense
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-04-02 Classification: OXIDOREDUCTASE Ligands: FAD, NA, CL, MLI |
 |
Organism: Phleum pratense
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-04-02 Classification: OXIDOREDUCTASE Ligands: FAD, NA, MLI |
 |
Organism: Phleum pratense
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-04-02 Classification: OXIDOREDUCTASE Ligands: FAD, NA, MLI |
 |
Organism: Phleum pratense
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2014-04-02 Classification: OXIDOREDUCTASE Ligands: FAD, NA, CL, MLI |
 |
Organism: Phleum pratense
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-04-02 Classification: OXIDOREDUCTASE Ligands: FAD, NA, MLI, XE |
 |
Organism: Phleum pratense
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-04-02 Classification: OXIDOREDUCTASE Ligands: FAD, NA, BR |

