Search Count: 23
All
Selected
 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dj85-B.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dj85-C.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dj85-D.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dj85-E.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Herh-C.01 Fab
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Trnm-B*01 Fab In Complex With Hiv-1 Env Trimer Bg505.Ds Sosip
Organism: Macaca mulatta, Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Herh-B*01 Fab In Complex With Hiv-1 Env Trimer Bg505.Ds Sosip
Organism: Human immunodeficiency virus 1, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Trnm-F*01 Fab In Complex With Hiv-1 Env Trimer Conc Sosip
Organism: Homo sapiens, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Hiv-1 Env Bg505 Ds-Sosip In Complex With Antibody Gpz6-B.01 Targeting The Fusion Peptide
Organism: Human immunodeficiency virus 1, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Herh-A.01 Fab
Organism: Macaca, Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: Viral Protein/Immune System Ligands: NAG |
 |
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Gpz6-A.01 Fab
Organism: Human immunodeficiency virus 1, Macaca
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: Viral PROTEIN/iMMUNE SYSTEM Ligands: NAG |
 |
Organism: Macaca mulatta, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2024-07-10 Classification: IMMUNE SYSTEM |
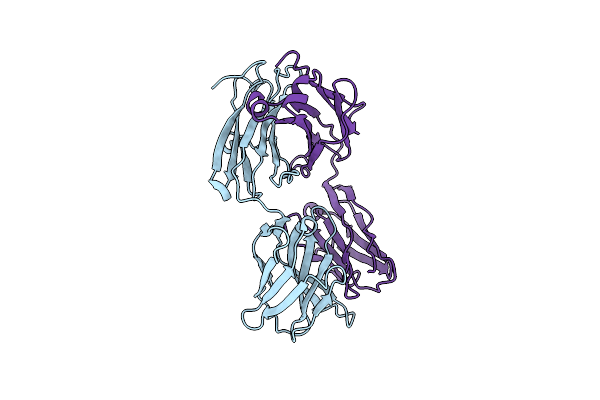 |
Organism: Macaca mulatta
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2024-07-10 Classification: IMMUNE SYSTEM |
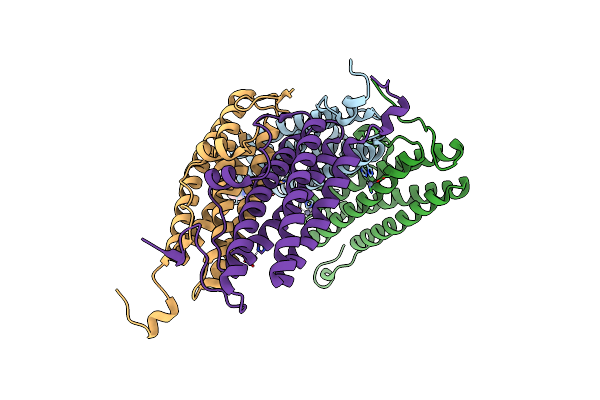 |
Organism: Nostoc punctiforme
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2017-02-01 Classification: METAL BINDING PROTEIN Ligands: ZN, EPE |
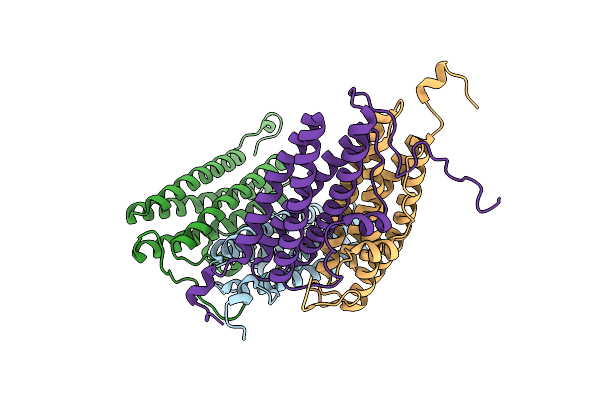 |
Organism: Nostoc punctiforme
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2017-01-25 Classification: METAL BINDING PROTEIN |
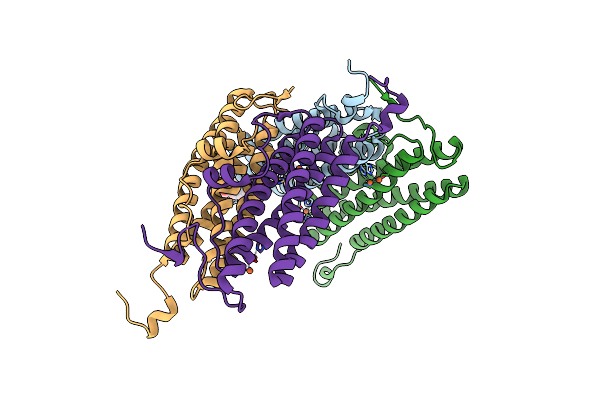 |
Organism: Nostoc punctiforme
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2017-01-25 Classification: METAL BINDING PROTEIN Ligands: FE, EPE |
 |
Structure Of The Imidazole-Adduct Of The Phormidium Laminosum Cytochrome C6 Q51V Variant
Organism: Phormidium laminosum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2011-02-09 Classification: PHOTOSYNTHESIS Ligands: HEC, IMD |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2007-07-24 Classification: PHOTOSYNTHESIS Ligands: HEC |
 |
Organism: Phormidium laminosum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-07-24 Classification: PHOTOSYNTHESIS Ligands: HEC, IMD, ZN, CL |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2006-07-05 Classification: ELECTRON TRANSFER Ligands: HEC |

