Search Count: 20
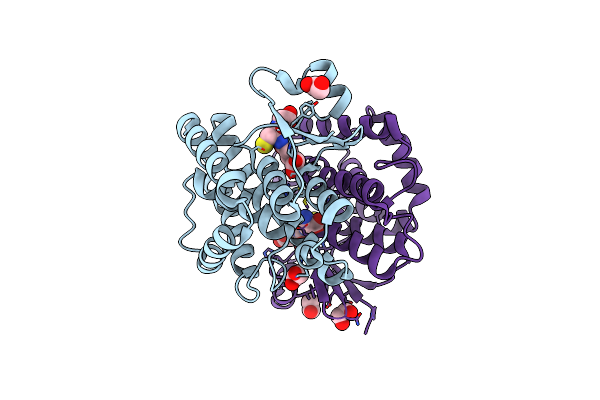 |
Structural Characterization Of Catalytic Site Of A Nilaparvata Lugens Delta-Class Glutathione Transferase
Organism: Nilaparvata lugens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-01-14 Classification: TRANSFERASE Ligands: GSH, EDO |
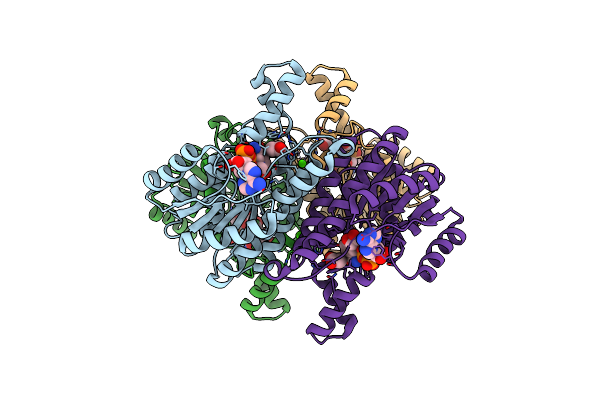 |
Crystal Structure Of Alcaligenes Faecalis D-3-Hydroxybutyrate Dehydrogenase In Complex With Nad(+) And Acetate
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-02-29 Classification: OXIDOREDUCTASE Ligands: ACT, CL, NAD, CA |
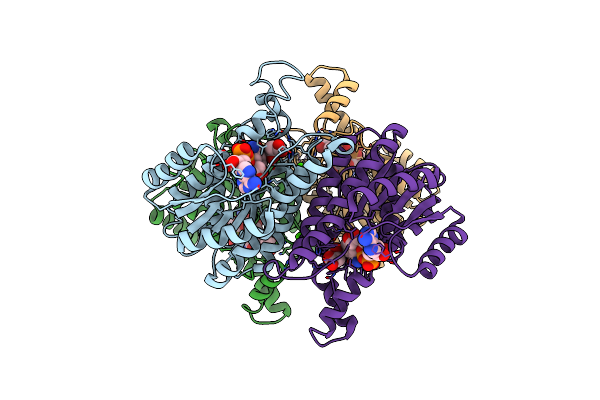 |
Crystal Structure Of D-3-Hydroxybutyrate Dehydrogenase, Prepared In The Presence Of The Substrate D-3-Hydroxybutyrate And Nad(+)
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-02-08 Classification: OXIDOREDUCTASE Ligands: CA, CL, NAD, NAI, 3HR, AAE |
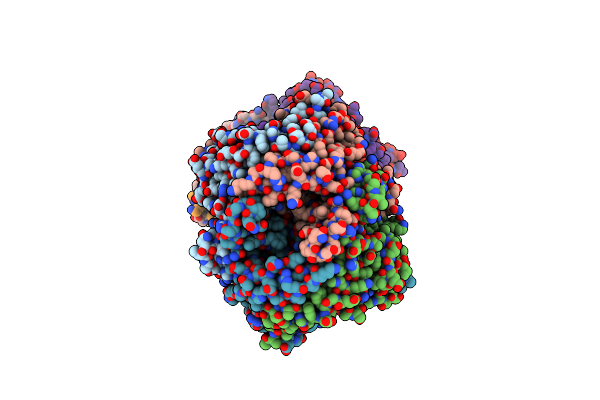 |
Crystal Structure Of Uricase From Arthrobacter Globiformis In Complex With Uric Acid (Substrate)
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Release Date: 2008-05-06 Classification: OXIDOREDUCTASE Ligands: URC |
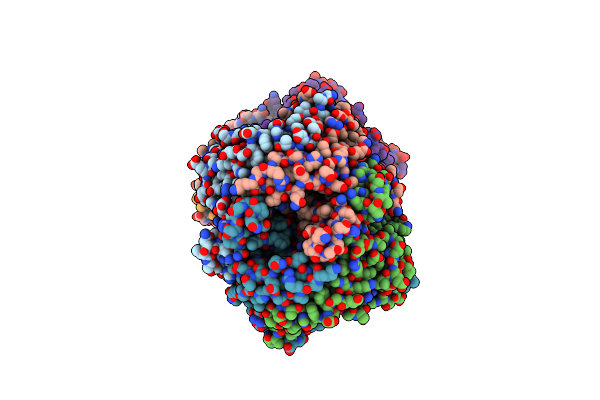 |
Crystal Structure Of Uricase From Arthrobacter Globiformis In Complex With Allantoate
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Release Date: 2008-05-06 Classification: OXIDOREDUCTASE Ligands: 1AL |
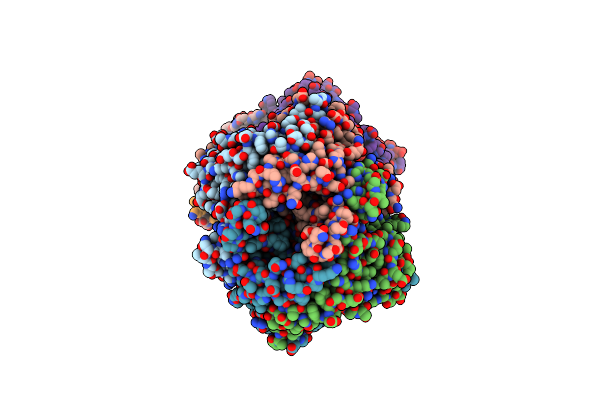 |
Crystal Structure Of Uricase From Arthrobacter Globiformis In Complex With 8-Azaxanthin (Inhibitor)
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Release Date: 2008-05-06 Classification: OXIDOREDUCTASE Ligands: AZA |
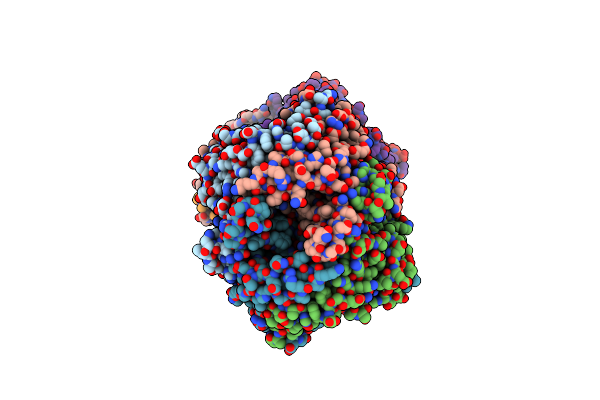 |
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Release Date: 2008-05-06 Classification: OXIDOREDUCTASE Ligands: NOB |
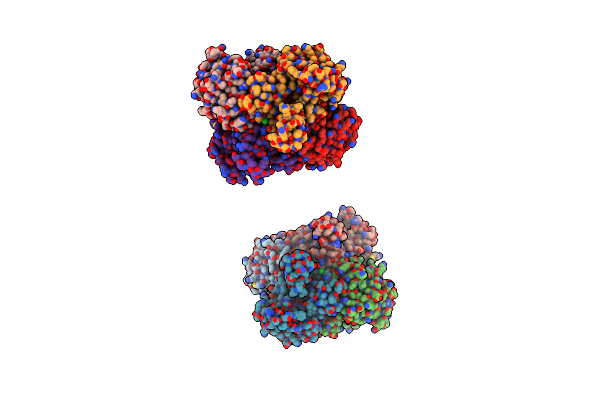 |
X-Ray Analyses Of 3-Hydroxybutyrate Dehydrogenase From Alcaligenes Faecalis
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2008-04-22 Classification: OXIDOREDUCTASE Ligands: CA, CL |
 |
Crystal Structures And Evolutionary Relationship Of Two Different Lipoamide Dehydrogenase(E3S) From Thermus Thermophilus
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-04-01 Classification: OXIDOREDUCTASE Ligands: CO3, FAD, PO4 |
 |
Crystal Structure Of Pyruvate Oxidase From Aerococcus Viridans Containing Fad
Organism: Aerococcus viridans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2007-05-29 Classification: OXIDOREDUCTASE Ligands: SO4, FAD |
 |
Organism: Sulfolobus tokodaii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-07-04 Classification: LIGASE Ligands: CL, SO4, EPE |
 |
Crystal Structure Of 2-Keto-3-Deoxygluconate Kinase (Form 1) From Sulfolobus Tokodaii
Organism: Sulfolobus tokodaii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2006-07-04 Classification: TRANSFERASE |
 |
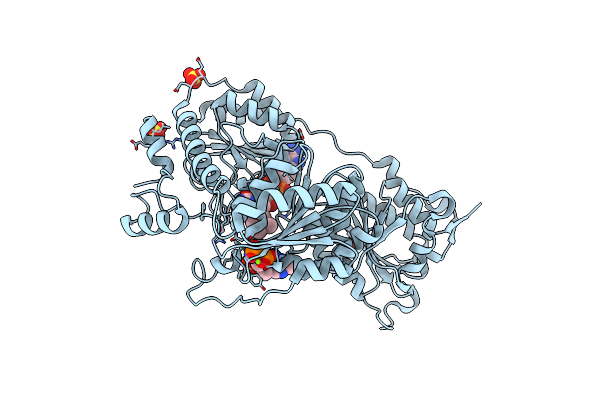
Crystal Structure Of Pyruvate Oxidase Containing Fad, From Aerococcus Viridans
Organism: Aerococcus viridans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2005-06-28 Classification: OXIDOREDUCTASE Ligands: SO4, FAD |
 |
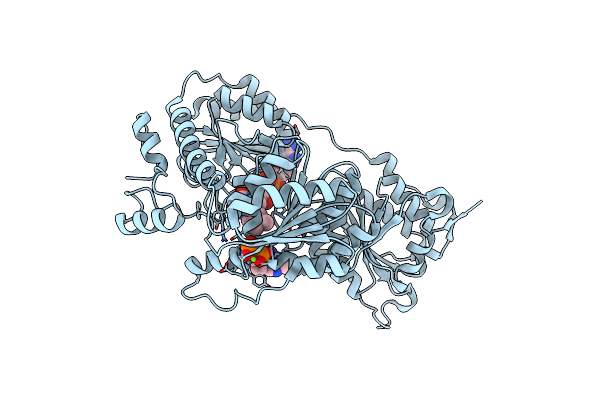
Crystal Structure Of Pyruvate Oxidase Complexed With Fad And Tpp, From Aerococcus Viridans
Organism: Aerococcus viridans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2005-06-28 Classification: OXIDOREDUCTASE Ligands: MG, SO4, FAD, TPP |
 |
Crystal Structure Of The Reaction Intermediate Between Pyruvate Oxidase Containing Fad And Tpp, And Substrate Pyruvate
Organism: Aerococcus viridans
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2005-06-28 Classification: OXIDOREDUCTASE Ligands: MG, FAD, HTL |
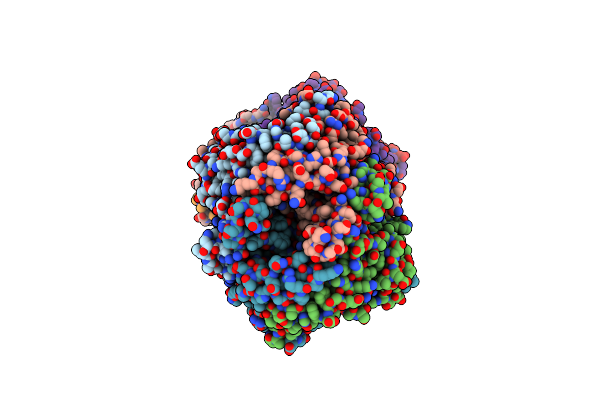 |
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2005-06-28 Classification: OXIDOREDUCTASE |
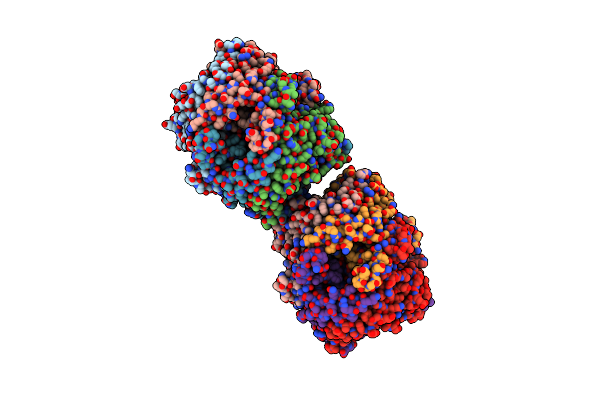 |
Crystal Structure Of Uricase From Arthrobacter Globiformis With Inhibitor 8-Azaxanthine
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2005-06-28 Classification: OXIDOREDUCTASE Ligands: AZA |
 |
Molecular And Crystal Structure Of D(Cgcgaatt(Mo4)Cgcg): The Watson-Crick Type And Wobble N4-Methoxycytidine/Guanosine Base Pairs In B-Dna
Method: X-RAY DIFFRACTION
Resolution:2.10 Å Release Date: 2003-02-11 Classification: DNA Ligands: MG |
 |
Molecular And Crystal Structure Of D(Cgcaaattmo4Cgcg): The Watson-Crick Type N4-Methoxycytidine/Adenosine Base Pair In B-Dna
Method: X-RAY DIFFRACTION
Resolution:1.60 Å Release Date: 2001-09-28 Classification: DNA Ligands: MG |
 |
Molecular And Crystal Structure Of D(Cgcgaatt(Mo4)Cgcg): The Watson-Crick Type And Wobble N4-Methoxycytidine/Guanosine Base Pairs In B-Dna
Method: X-RAY DIFFRACTION
Resolution:1.60 Å Release Date: 2001-09-14 Classification: DNA Ligands: MG |

