Search Count: 67
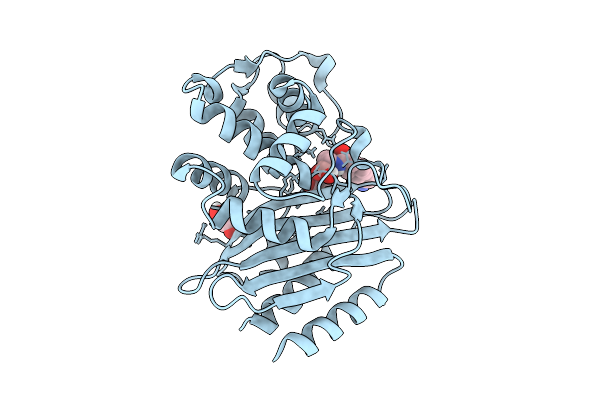 |
Crystal Structure Of Peni Beta-Lactamase From Burkholderia Pseudomallei Complex With Taniborbactam
Organism: Burkholderia pseudomallei k96243
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: HYDROLASE Ligands: KJK, GOL |
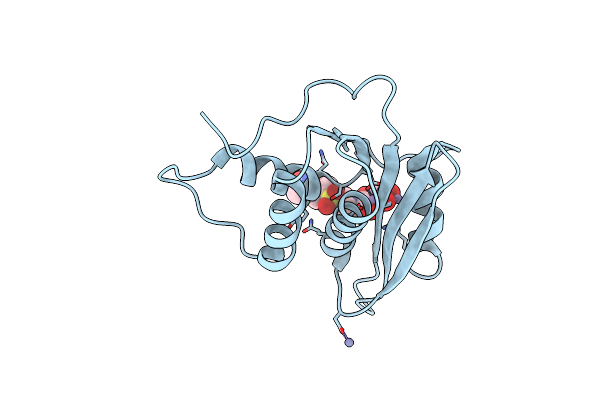 |
Crystal Structure Of Hiv-1 Reverse Transcriptase Rnase H Domain Complexed With A Galloyl Inhibitor
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: VIRAL PROTEIN Ligands: MN, ZN, A1L9C |
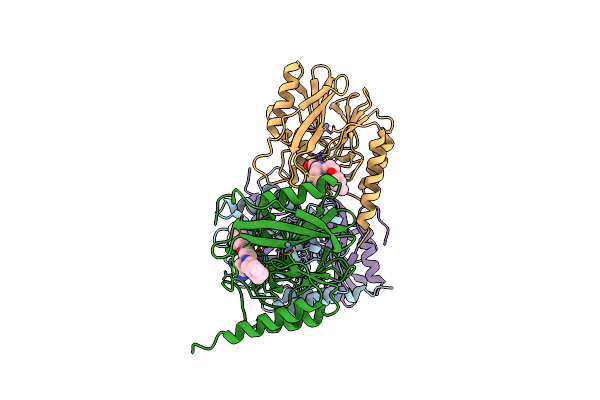 |
Crystal Structure Of Metallo-Beta-Lactamse, Imp-1, Complexed With A Quinolinone-Based Inhibitor
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-05-08 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, A1LXO |
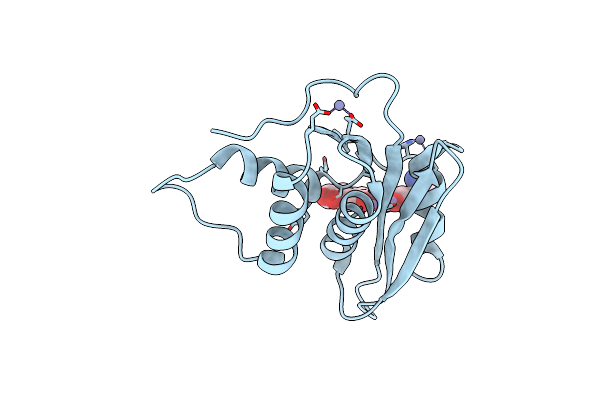 |
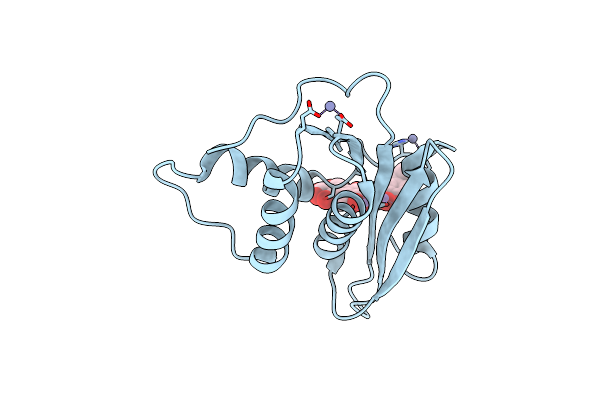
Crystal Structure Of Engineered Hiv-1 Reverse Transcriptase Rnase H Domain Complexed With Laccaic Acid C
Organism: Hiv-1 06tg.ht008
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2023-08-16 Classification: VIRAL PROTEIN Ligands: MN, V96, ZN |
 |
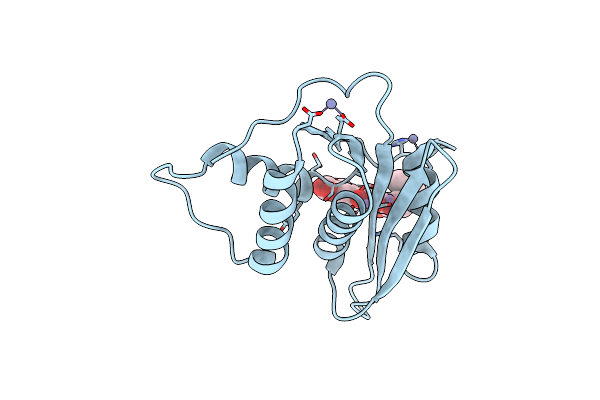
Crystal Structure Of Engineered Hiv-1 Reverse Transcriptase Rnase H Domain Complexed With Laccaic Acid E
Organism: Hiv-1 06tg.ht008
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2023-08-16 Classification: VIRAL PROTEIN Ligands: MN, V9O, ZN |
 |
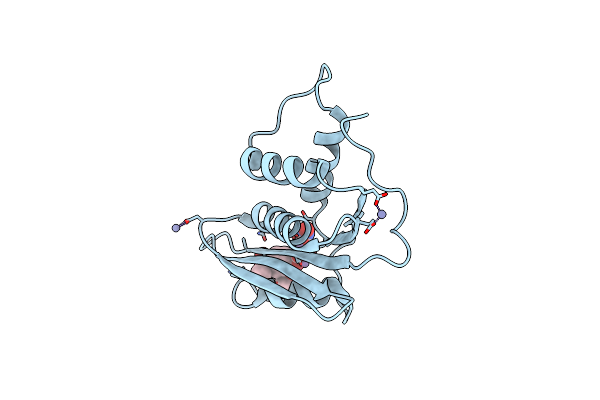
Crystal Structure Of Engineered Hiv-1 Reverse Transcriptase Rnase H Domain Complexed With Laccaic Acid A
Organism: Hiv-1 06tg.ht008
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2023-08-16 Classification: VIRAL PROTEIN Ligands: MN, VA9, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2023-03-08 Classification: CELL CYCLE |
 |
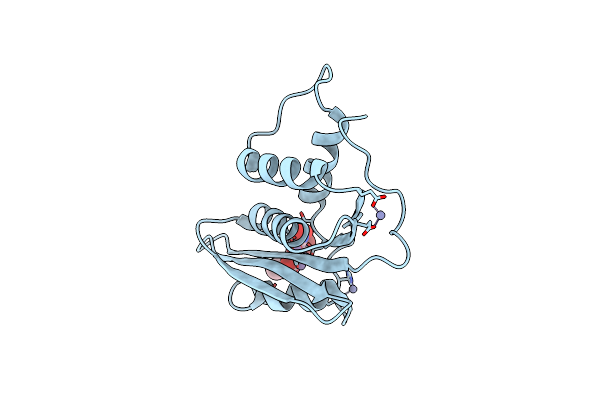
Crystal Structure Of Engineered Hiv-1 Reverse Transcriptase Rnase H Domain Complexed With Nitrofuran Methoxy(Methoxycarbonyl)Phenyl Ester
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2022-04-27 Classification: VIRAL PROTEIN Ligands: MN, E58, ZN |
 |
Crystal Structure Of Engineered Hiv-1 Reverse Transcriptase Rnase H Domain Complexed With Nitrofuran Methoxy(Methoxycarbonyl)Phenyl Ester
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2022-04-27 Classification: VIRAL PROTEIN Ligands: MN, ZN, E6I |
 |
Crystal Structure Of Engineered Hiv-1 Reverse Transcriptase Rnase H Domain Complexed With Nitrofuran Methoxy(Methoxycarbonyl)Phenyl Ester
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2022-04-27 Classification: VIRAL PROTEIN Ligands: MN, E81, ZN |
 |
Crystal Structure Of Engineered Hiv-1 Reverse Transcriptase Rnase H Domain Complexed With Nitrofuran Methoxy(Methoxycarbonyl)Phenyl Ester
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-04-27 Classification: VIRAL PROTEIN Ligands: MN, ZN, E9B |
 |
Crystal Structure Of Engineered Hiv-1 Reverse Transcriptase Rnase H Domain Complexed With Nitrofuran Methoxy(Methoxycarbonyl)Phenyl Ester
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-04-27 Classification: VIRAL PROTEIN Ligands: ECW, MN, ZN |
 |
Crystal Structure Of Engineered Hiv-1 Reverse Transcriptase Rnase H Domain Complexed With Nitrofuran Methoxy(Methoxycarbonyl)Phenyl Ester
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-04-27 Classification: VIRAL PROTEIN Ligands: MN, ZN, EGI |
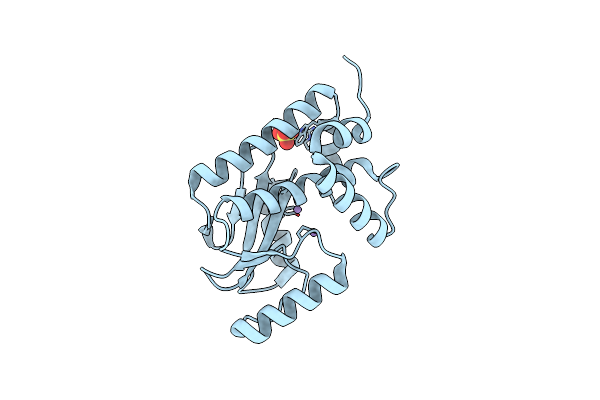 |
Crystal Structure Of Influenza Polymerase Acidic Subunit N-Terminal Domain Crystallized By Ammonium Sulfate With Glycan
Organism: Influenza a virus (a/puerto rico/8/1934(h1n1))
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-04-20 Classification: VIRAL PROTEIN Ligands: SO4, MN |
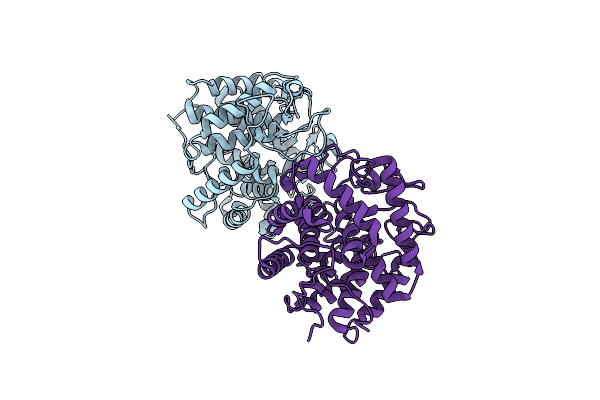 |
Crystal Structure Of Chitosanase Crystallized By Ammonium Sulfate With Glycan
Organism: Bacillus sp. k17-2
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-04-20 Classification: HYDROLASE |
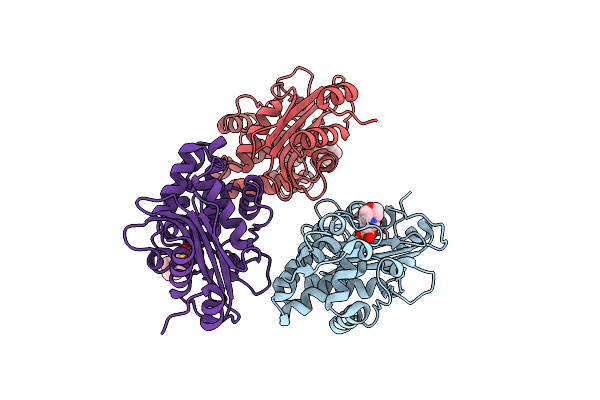 |
Crystal Structure Of R220A Variant Pena Beta-Lactamase From Burkholderia Multivorans
Organism: Burkholderia multivorans (strain atcc 17616 / 249)
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2021-03-17 Classification: HYDROLASE Ligands: MES |
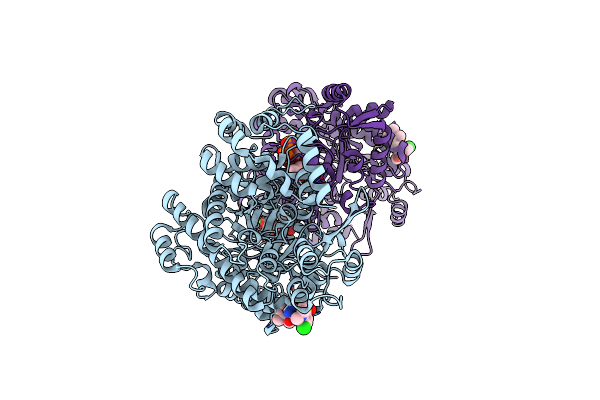 |
Crystal Structure Of Ribonucleotide Reductase R1 Subunit, Rrm1 In Complex With 5-Chloro-N-((1S,2R)-2-(6-Fluoro-2,3-Dimethylphenyl)-1-(5-Oxo-4,5-Dihydro-1,3,4-Oxadiazol-2-Yl)Propyl)-4-Methyl-3,4-Dihydro-2H-Benzo[B][1,4]Oxazine-8-Sulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2020-12-23 Classification: OXIDOREDUCTASE Ligands: MG, TTP, EJ6 |
 |
Crystal Structure Of Ribonucleotide Reductase R1 Subunit, Rrm1 In Complex With 5-Chloro-2-(N-((1S,2R)-2-(2,3-Dihydro-1H-Inden-4-Yl)-1-(5-Oxo-4,5-Dihydro-1,3,4-Oxadiazol-2-Yl)Propyl)Sulfamoyl)Benzamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2020-11-04 Classification: OXIDOREDUCTASE Ligands: MG, TTP, E6O, ACT |
 |
Crystal Structure Of Ribonucleotide Reductase R1 Subunit, Rrm1 In Complex With 4-Bromo-N-((1S,2R)-2-(Naphthalen-1-Yl)-1-(5-Oxo-4,5-Dihydro-1,3,4-Oxadiazol-2-Yl)Propyl)Benzenesulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-10-28 Classification: OXIDOREDUCTASE Ligands: TTP, MG, E4X, ACT, SO4 |
 |
Crystal Structure Of Inactive Form Of Chitosanase Crystallized By Ammonium Sulfate
Organism: Bacillus sp. k17-2
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2020-07-29 Classification: HYDROLASE |

