Search Count: 16
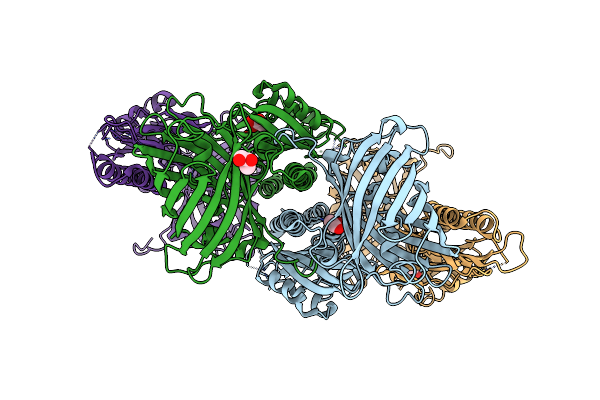 |
Chimeric Fluorescence Biosensor Formed From A Lactate-Binding Protein And Gfp, Bound To Lactate
Organism: Aequorea victoria, Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2025-03-19 Classification: FLUORESCENT PROTEIN Ligands: GOL, LAC |
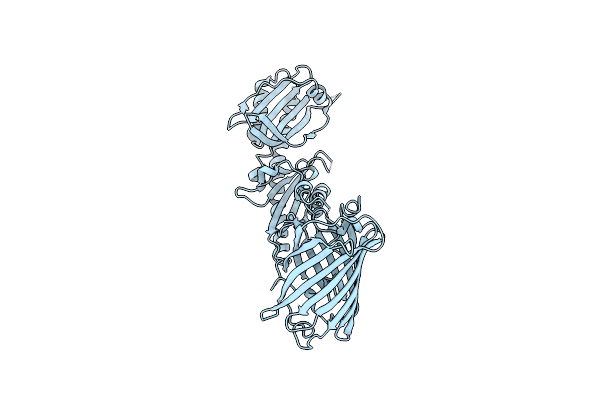 |
Chimeric Fluorescence Biosensor Formed From A Lactate-Binding Protein And Gfp
Organism: Aequorea victoria, Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2025-03-19 Classification: FLUORESCENT PROTEIN |
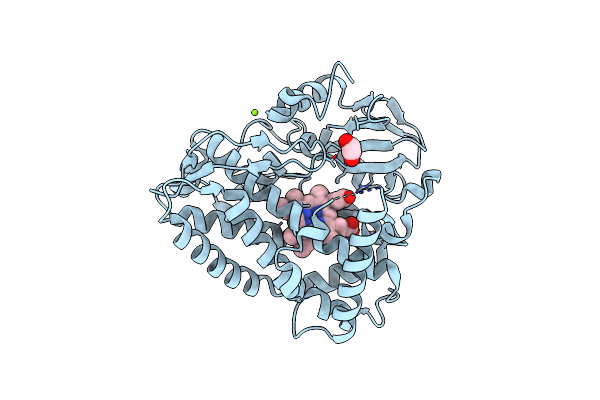 |
Organism: Streptomyces atratus
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2023-12-13 Classification: METAL BINDING PROTEIN Ligands: HEM, MG, CL, EDO |
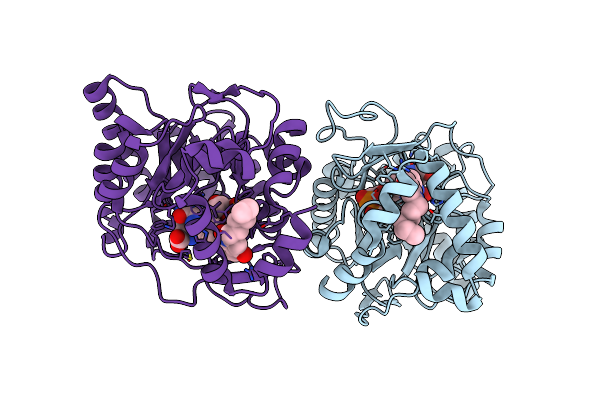 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-03-02 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: FMN, HQO, ORO |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2022-03-02 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: F2D, PO4, ORO, FMN |
 |
E. Coli Dihydroorotate Dehydrogenase Bound To The Ubiquinone Surrogate Dcip
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2022-03-02 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: PE4, ORO, RLM, FMN, GOL |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2022-03-02 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: ORO, FMN |
 |
Organism: Rattus norvegicus, Gallus gallus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-08 Classification: CELL CYCLE Ligands: GTP, MG, GOL, GDP, L95, MES, ACP |
 |
Crystal Structure Of Human Cellular Retinol Binding Protein 1 In Complex With Abnormal-Cannabidiol (Abn-Cbd)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2019-02-13 Classification: LIPID BINDING PROTEIN Ligands: HVD |
 |
Crystal Structure Of Human Cellular Retinol Binding Protein 1 In Complex With Abnormal-Cannabidiorcin (Abn-Cbdo)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-02-13 Classification: LIPID BINDING PROTEIN Ligands: HVJ |
 |
Crystal Structure Of Human Cellular Retinol Binding Protein 3 In Complex With Abnormal-Cannabidiol (Abn-Cbd)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-02-13 Classification: LIPID BINDING PROTEIN Ligands: HVD, GOL |
 |
Crystal Structure Of Human Cellular Retinol-Binding Protein 4 In Complex With Abnormal-Cannabidiol (Abn-Cbd)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-02-13 Classification: LIPID BINDING PROTEIN Ligands: HVD |
 |
Crystal Structure Of Human Cellular Retinol-Binding Protein 1 In Complex With Cannabidiorcin (Cbdo)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-02-13 Classification: LIPID BINDING PROTEIN Ligands: 8CB |
 |
Organism: Rattus norvegicus, Gallus gallus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-09-20 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, 7AO, MES, ACP |
 |
Organism: Rattus norvegicus, Gallus gallus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-03-01 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, MES, 7AK, ACP |
 |
Organism: Rattus norvegicus, Gallus gallus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2016-06-29 Classification: CELL CYCLE Ligands: GTP, MG, CA, IMD, GOL, GDP, 6K9, 03S, MES, ACP |

