Search Count: 23
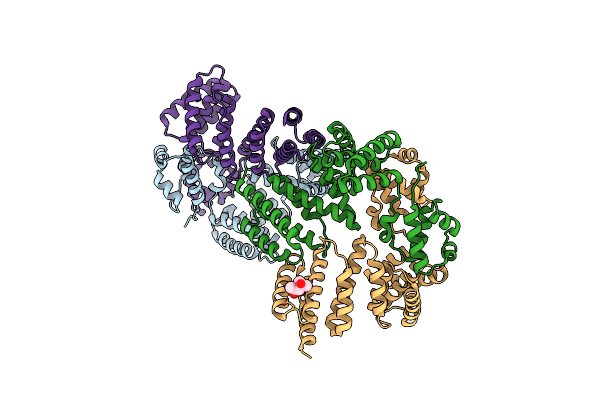 |
Crystal Structure Of Leucine-Rich Protein Regulator, Elrr, From Enterococcus Faecalis
Organism: Enterococcus faecalis (strain atcc 700802 / v583)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-09-19 Classification: TRANSCRIPTION Ligands: MPD |
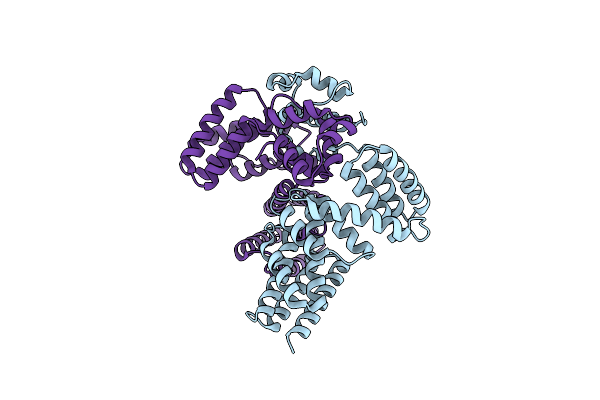 |
Crystal Structure Of Leucine-Rich Protein Regulator, Elrr, From Enterococcus Faecalis
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2018-09-19 Classification: TRANSCRIPTION |
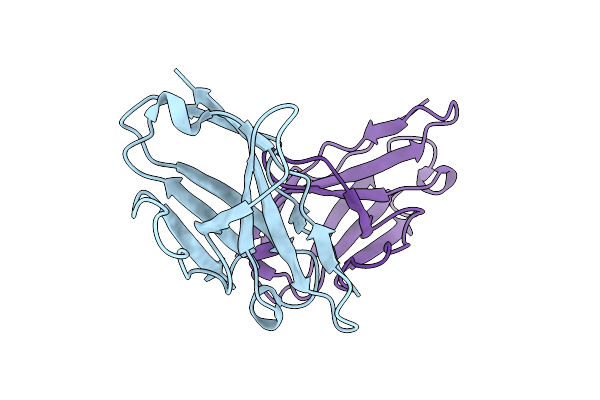 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2009-11-17 Classification: IMMUNE SYSTEM |
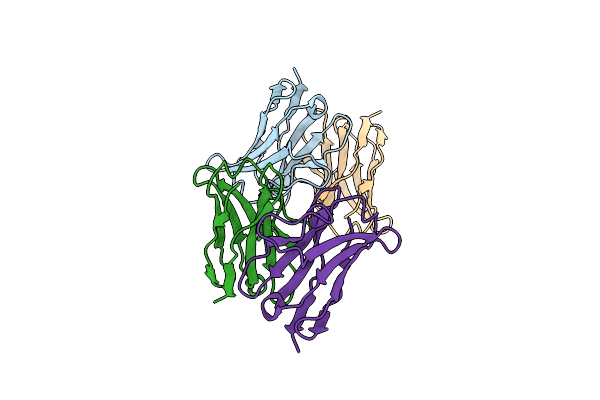 |
Crystal Structure Of The Mutant H8P From The Recombinant Variable Domain 6Jal2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-11-17 Classification: IMMUNE SYSTEM |
 |
Glucosamine-6-Phosphate Deaminase Complexed With The Allosteric Activator N-Acetyl-Glucoamine-6-Phosphate Both In The Active And Allosteric Sites.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-10-06 Classification: HYDROLASE Ligands: 16G, FGS |
 |
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-09-15 Classification: ISOMERASE Ligands: SO4 |
 |
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2009-09-15 Classification: ISOMERASE Ligands: SO4, AZI |
 |
Crystallographic Structure Of Betaine Aldehyde Dehydrogenase From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-08-04 Classification: OXIDOREDUCTASE Ligands: NAP, BME, K, GOL |
 |
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-01-13 Classification: OXIDOREDUCTASE Ligands: NAG, HEM |
 |
Crystal Structure Of The Unstable And Highly Fibrillogenic Pro7Ser Mutant Of The Recombinant Variable Domain 6Ajl2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-11-04 Classification: IMMUNE SYSTEM Ligands: ACT, GOL, NA |
 |
Crystal Structure Of The Unstable And Highly Fibrillogenic Pro7Ser Mutant Of The Recombinant Variable Domain 6Ajl2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-10-28 Classification: IMMUNE SYSTEM Ligands: ACT, GOL, MES |
 |
Crystal Structure Of The Catalase-1 From Neurospora Crassa, Native Structure At 1.75A Resolution.
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2004-10-12 Classification: OXIDOREDUCTASE Ligands: HDD, HEM |
 |
Human Glucosamine-6-Phosphate Deaminase Isomerase At 1.75 A Resolution Complexed With N-Acetyl-Glucosamine-6-Phosphate And 2-Deoxy-2-Amino-Glucitol-6-Phosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2003-09-23 Classification: HYDROLASE Ligands: 16G, SO4, AGP |
 |
Structure Of The Mutant F174A T Form Of The Glucosamine-6-Phosphate Deaminase From E.Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2002-02-20 Classification: HYDROLASE |
 |
Glucosamine 6-Phosphate Deaminase Complexed With The Substrate Of The Reverse Reaction Fructose 6-Phosphate (Open Form)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2002-01-04 Classification: ISOMERASE Ligands: F6R |
 |
Glucosamine-6-Phosphate Deaminase From E.Coli, R Conformer. Complexed With The Allosteric Activator N-Acetyl-Glucosamine-6-Phosphate At 2.2 A Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2002-01-04 Classification: ISOMERASE Ligands: 16G |
 |
A Discovery Of Three Alternate Conformations In The Active Site Of Glucosamine-6-Phosphate Isomerase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2002-01-04 Classification: ISOMERASE Ligands: 16G, TLA |
 |
Glucosamine-6-Phosphate Deaminase From E.Coli, T Conformer, At 2.2A Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2002-01-04 Classification: ISOMERASE |
 |
Glucosamine-6-Phosphate Deaminase From E.Coli, T Conformer, At 1.9A Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2002-01-04 Classification: ISOMERASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2000-03-06 Classification: ISOMERASE |

