Search Count: 14
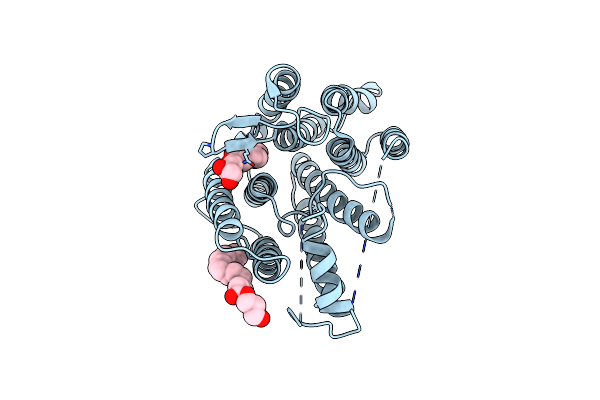 |
Crystal Structure Of Thermus Thermophilus Rod Shape Determining Protein Roda (Q5Six3_Thet8)
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2018-03-28 Classification: TRANSFERASE Ligands: OLC, CL |
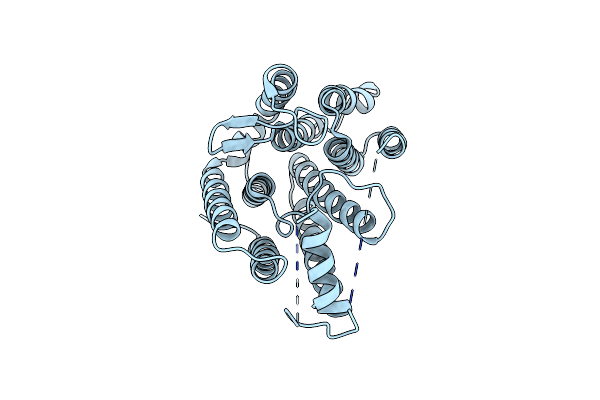 |
Crystal Structure Of Thermus Thermophilus Rod Shape Determining Protein Roda D255A Mutant (Q5Six3_Thet8)
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2018-03-28 Classification: TRANSFERASE Ligands: CL |
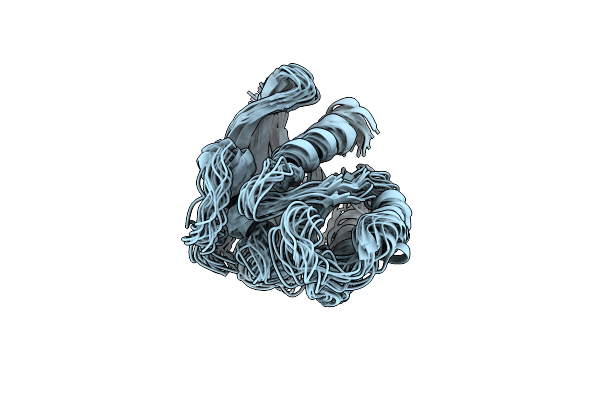 |
Ec-Nmr Structure Of Human H-Rast35S Mutant Protein Determined By Combining Evolutionary Couplings (Ec) And Sparse Nmr Data
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2015-07-01 Classification: SIGNALING PROTEIN |
 |
Ec-Nmr Structure Of Escherichia Coli Maltose-Binding Protein Determined By Combining Evolutionary Couplings (Ec) And Sparse Nmr Data. Northeast Structural Genomics Consortium Target Er690
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2015-07-01 Classification: PERIPLASMIC BINDING PROTEIN |
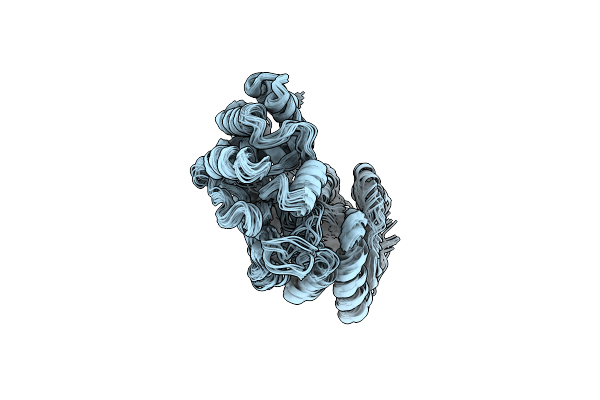 |
Ec-Nmr Structure Of Escherichia Coli Maltose-Binding Protein Determined By Combining Evolutionary Couplings (Ec) And Sparse Nmr Data With A Second Set Of Rdc Data Simulated For An Alternative Alignment Tensor. Northeast Structural Genomics Consortium Target Er690
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2015-07-01 Classification: PERIPLASMIC BINDING PROTEIN |
 |
Ec-Nmr Structure Of Human H-Rast35S Mutant Protein Determined By Combining Evolutionary Couplings (Ec) And Sparse Nmr Data
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2015-07-01 Classification: SIGNALING PROTEIN |
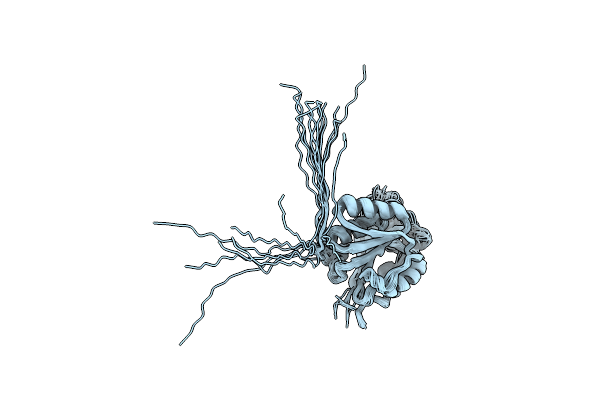 |
Ec-Nmr Structure Of Synechocystis Sp. Pcc 6803 Slr1183 Determined By Combining Evolutionary Couplings (Ec) And Sparse Nmr Data. Northeast Structural Genomics Consortium Target Sgr145
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: SOLUTION NMR Release Date: 2015-07-01 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
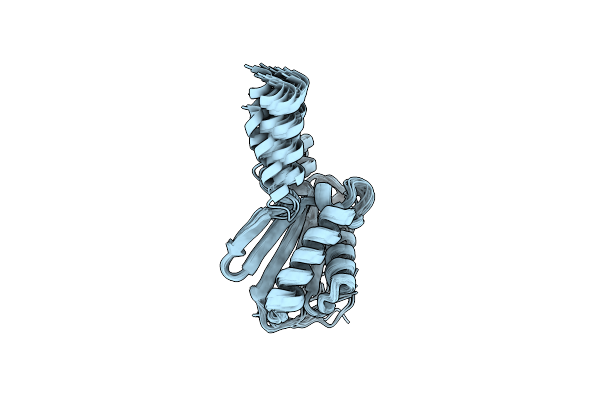 |
Ec-Nmr Structure Of Escherichia Coli Yiad Determined By Combining Evolutionary Couplings (Ec) And Sparse Nmr Data. Northeast Structural Genomics Consortium Target Er553
Organism: Escherichia coli k-12
Method: SOLUTION NMR Release Date: 2015-07-01 Classification: LIPID BINDING PROTEIN |
 |
Ec-Nmr Structure Of Erwinia Carotovora Eca1580 N-Terminal Domain Determined By Combining Evolutionary Couplings (Ec) And Sparse Nmr Data. Northeast Structural Genomics Consortium Target Ewr156A
Organism: Pectobacterium atrosepticum scri1043
Method: SOLUTION NMR Release Date: 2015-07-01 Classification: Structural Genomics, Unknown Function |
 |
Ec-Nmr Structure Of Ralstonia Metallidurans Rmet_5065 Determined By Combining Evolutionary Couplings (Ec) And Sparse Nmr Data. Northeast Structural Genomics Consortium Target Crr115
Organism: Cupriavidus metallidurans ch34
Method: SOLUTION NMR Release Date: 2015-07-01 Classification: Structural Genomics, Unknown Function |
 |
Ec-Nmr Structure Of Ralstonia Metallidurans Rmet_5065 Determined By Combining Evolutionary Couplings (Ec) And Sparse Nmr Data. Northeast Structural Genomics Consortium Target Crr115
Organism: Cupriavidus metallidurans ch34
Method: SOLUTION NMR Release Date: 2015-07-01 Classification: Structural Genomics, Unknown Function |
 |
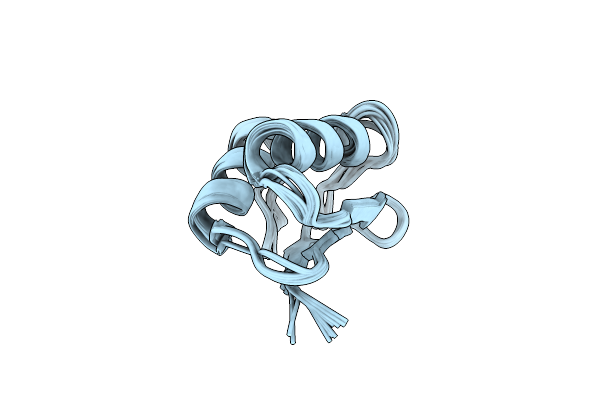
Ec-Nmr Structure Of Agrobacterium Tumefaciens Atu1203 Determined By Combining Evolutionary Couplings (Ec) And Sparse Nmr Data. Northeast Structural Genomics Consortium Target Att10
Organism: Agrobacterium fabrum str. c58
Method: SOLUTION NMR Release Date: 2015-07-01 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
 |
Ec-Nmr Structure Of Agrobacterium Tumefaciens Atu1203 Determined By Combining Evolutionary Couplings (Ec) And Sparse Nmr Data. Northeast Structural Genomics Consortium Target Att10
Organism: Agrobacterium fabrum str. c58
Method: SOLUTION NMR Release Date: 2015-07-01 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
 |
Ec-Nmr Structure Of Arabidopsis Thaliana At2G32350 Determined By Combining Evolutionary Couplings (Ec) And Sparse Nmr Data. Northeast Structural Genomics Consortium Target Ar3433A
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2015-07-01 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |

