Search Count: 119
 |
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-05-14 Classification: REPLICATION |
 |
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-07-26 Classification: DNA BINDING PROTEIN Ligands: FMT, ACT |
 |
Organism: Mycolicibacterium thermoresistibile atcc 19527, Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-01-18 Classification: DNA BINDING PROTEIN Ligands: ACE, NH2 |
 |
Organism: Mycolicibacterium thermoresistibile, Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-01-18 Classification: DNA BINDING PROTEIN Ligands: SO4, CL, ACE, NH2 |
 |
Organism: Mycolicibacterium thermoresistibile atcc 19527
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2023-01-18 Classification: DNA BINDING PROTEIN |
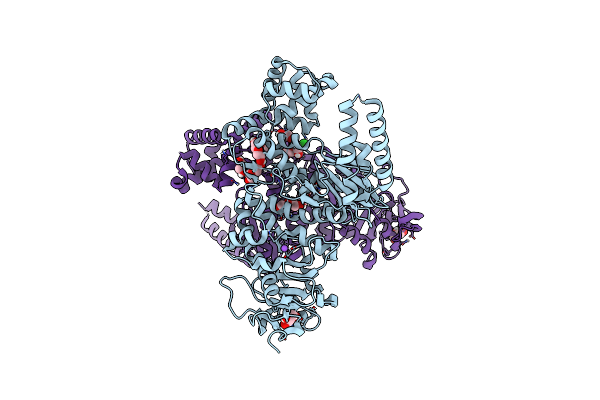 |
Plasmodium Falciparum Apicoplast Dna Polymerase (Exo-Minus) Without Affinity Tag
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-10-26 Classification: REPLICATION, Transferase Ligands: GOL, NA, CL, PEG, EDO |
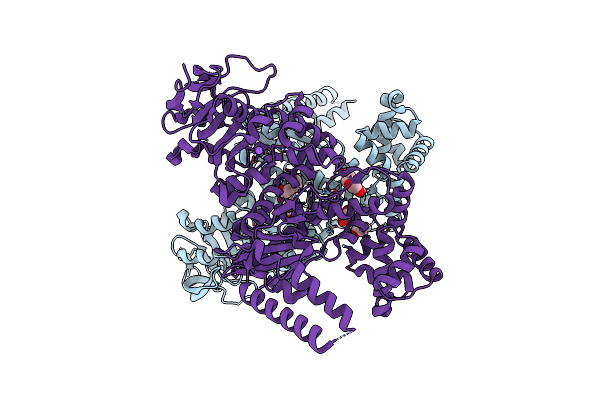 |
Plasmodium Falciparum Apicoplast Dna Polymerase (Exo-Minus) Without Affinity Tag
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-10-19 Classification: REPLICATION, TRANSFERASE Ligands: PEG, CL, NA, EDO |
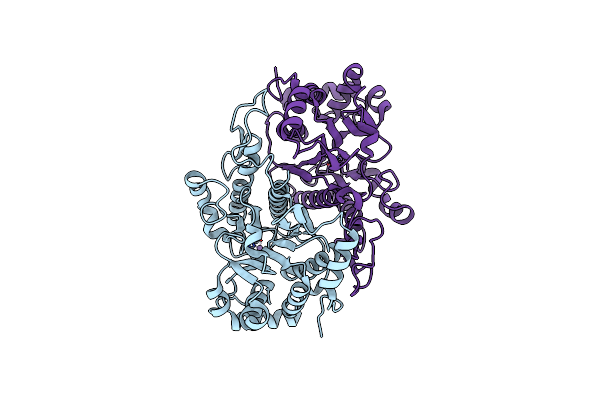 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-05-23 Classification: TRANSFERASE Ligands: MN |
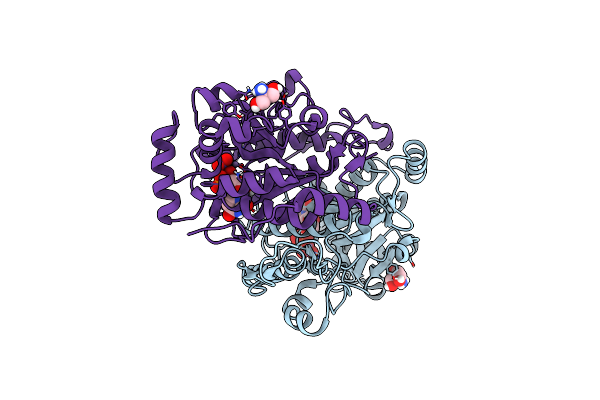 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2018-05-23 Classification: TRANSFERASE Ligands: MN, UDP, TRS, NO3 |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2018-05-23 Classification: TRANSFERASE Ligands: MN, UDP, GOL, TRS, NO3 |
 |
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-08-10 Classification: TRANSFERASE Ligands: SO4, NA |
 |
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-08-10 Classification: TRANSFERASE Ligands: MG |
 |
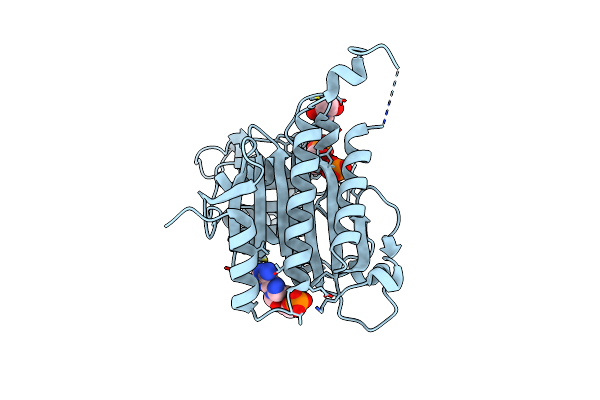
Product Complexes Of Porcine Liver Fructose-1,6-Bisphosphatase With Mutation E192Q
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-09-18 Classification: HYDROLASE Ligands: F6P, ZN, PO4 |
 |
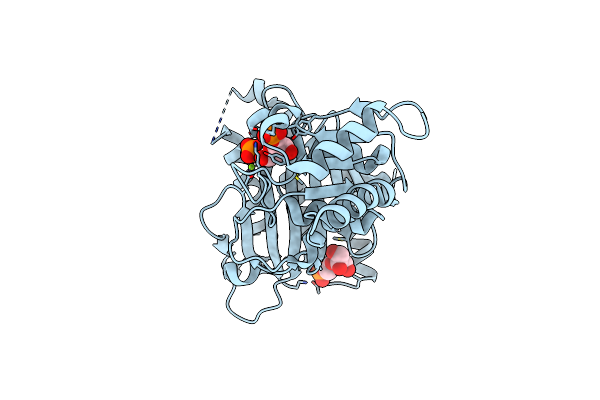
Crystal Structure Of Amp Complexes Of Nem Modified Porcine Liver Fructose-1,6-Bisphosphatase
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-09-18 Classification: HYDROLASE Ligands: F6P, MG, PO4, AMP, NEN |
 |
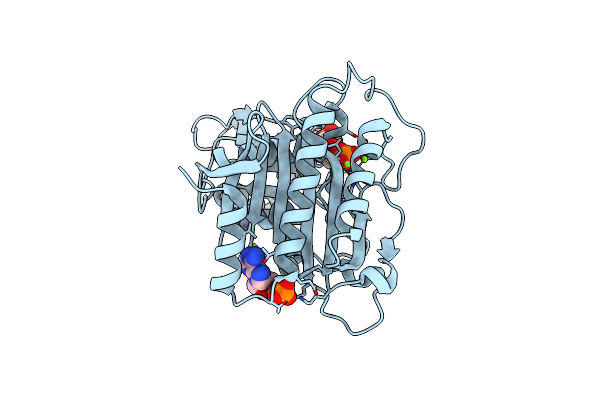
Crystal Structure Of Product Complexes Of Porcine Liver Fructose-1,6-Bisphosphatase With Blocked Subunit Pair Rotation
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2013-09-11 Classification: HYDROLASE Ligands: F6P, MG, PO4 |
 |
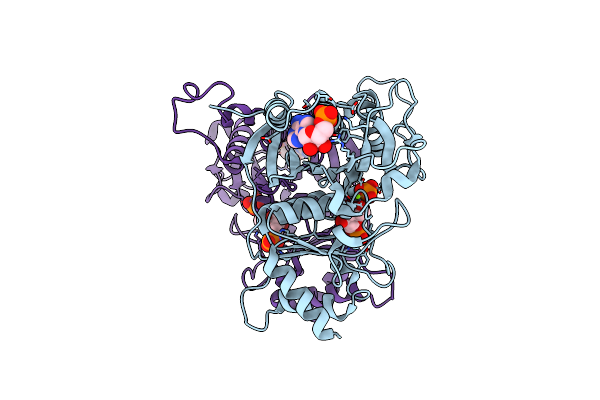
Crystal Structure Of Amp Complexes Of Porcine Liver Fructose-1,6-Bisphosphatase With Blocked Subunit Pair Rotation
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2013-09-11 Classification: HYDROLASE Ligands: F6P, PO4, MG, AMP |
 |
Crystal Structure Of Amp Complexes Of Porcine Liver Fructose-1,6-Bisphosphatase With Filled Central Cavity
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2013-09-04 Classification: HYDROLASE Ligands: F6P, MG, AMP, PO4 |
 |
Crystal Structure Of Fru 2,6-Bisphosphate Complexes Of Porcine Liver Fructose-1,6-Bisphosphatase With Filled Central Cavity
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2013-09-04 Classification: HYDROLASE Ligands: FDP, MG |
 |
Crystal Structure Of Product Complexes Of Porcine Liver Fructose-1,6-Bisphosphatase With Restrained Subunit Pair Rotation
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2013-09-04 Classification: HYDROLASE Ligands: F6P, PO4, MG |
 |
Crystal Structure Of Amp Complexes Of Porcine Liver Fructose-1,6-Bisphosphatase With Restrained Subunit Pair Rotation
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-09-04 Classification: HYDROLASE Ligands: F6P, MG, AMP, PO4 |

