Search Count: 105
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: PROTEIN TRANSPORT Ligands: CO3, NAG, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: E1J, H81 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: VIRAL PROTEIN, HYDROLASE Ligands: WZK |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: VIRAL PROTEIN, HYDROLASE |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: STRUCTURAL PROTEIN |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: STRUCTURAL PROTEIN |
 |
Organism: Autographa californica nucleopolyhedrovirus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: STRUCTURAL PROTEIN Ligands: NAG |
 |
Organism: Autographa californica nucleopolyhedrovirus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: STRUCTURAL PROTEIN Ligands: NAG |
 |
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2024-07-31 Classification: HYDROLASE/DNA Ligands: ADP, ALF, MG, K |
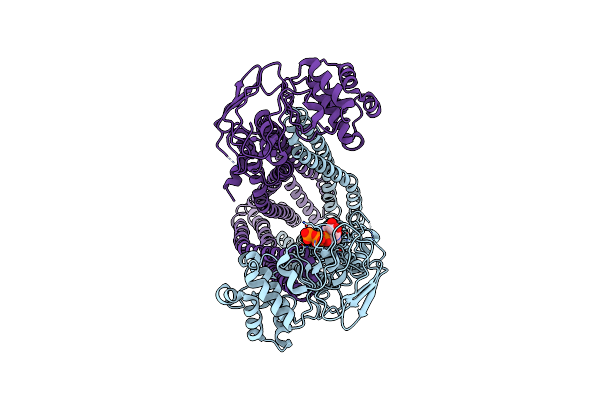 |
Cryo-Em Structure Of Human Abcd1 E630Q In The Presence Of Atp In Inward-Facing State 2
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: STRUCTURAL PROTEIN Ligands: ATP |
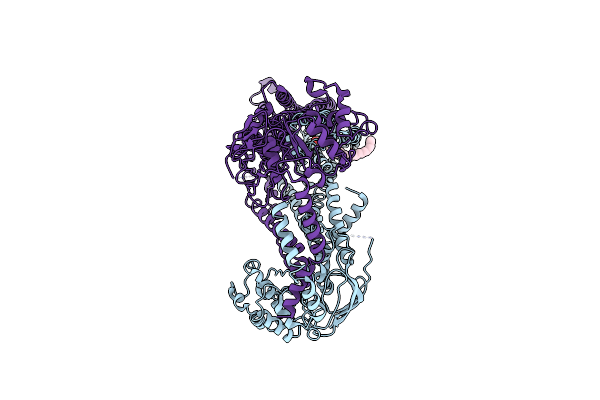 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: STRUCTURAL PROTEIN Ligands: 7PO |
 |
Cryo-Em Structure Of Human Abcd1 E630Q In The Presence Of C26:0-Coa And Atp
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: STRUCTURAL PROTEIN Ligands: CO8, ATP |
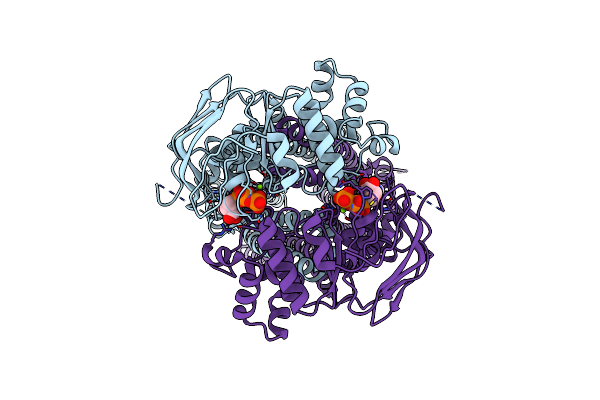 |
Cryo-Em Structure Of Human Abcd1 E630Q In The Presence Of Atp And Magnesium In Outward-Facing State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: STRUCTURAL PROTEIN Ligands: MG, ATP |
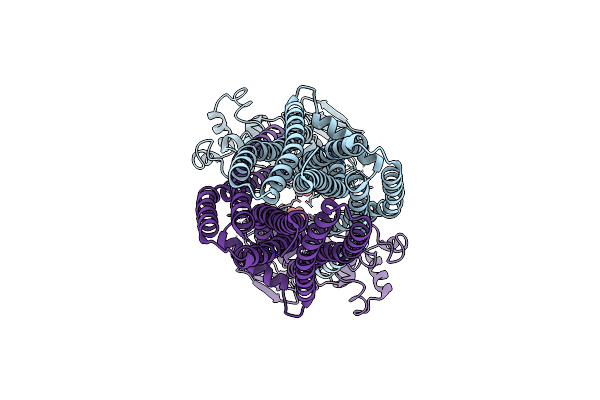 |
Cryo-Em Structure Of Human Abcd1 E630Q In The Presence Of Atp In Inward-Facing State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: STRUCTURAL PROTEIN Ligands: ATP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: STRUCTURAL PROTEIN Ligands: K9G |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2023-01-25 Classification: LIPID BINDING PROTEIN Ligands: KO0 |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-01-25 Classification: LIPID BINDING PROTEIN Ligands: IUF |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-01-25 Classification: LIPID BINDING PROTEIN Ligands: IUJ |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2023-01-25 Classification: LIPID BINDING PROTEIN Ligands: IUO, PO4 |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2023-01-25 Classification: LIPID BINDING PROTEIN Ligands: IUC |

