Search Count: 173
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: TRANSCRIPTION Ligands: A1D7W |
 |
Organism: Echinococcus multilocularis
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: E1J, H81 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2025-04-23 Classification: TRANSCRIPTION Ligands: GOL, A1D72 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-02-12 Classification: TRANSFERASE Ligands: A1A79, EDO |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.52 Å Release Date: 2025-01-22 Classification: ELECTRON TRANSPORT Ligands: LOP, CDL, HEM, A1IKG, LMT, HEC, FES, PSC, SO4 |
 |
Solution Nmr Structure Of Halichondamide A, A Fused Bicyclic Cysteine Knot Undecapeptide From The Marine Sponge Halichondria Bowerbanki
Organism: Halichondria bowerbanki
Method: SOLUTION NMR Release Date: 2024-08-21 Classification: BIOSYNTHETIC PROTEIN |
 |
Co-Crystal Structure Of Sos-1 And A Potent, Selective And Orally Bioavailable Sos1 Inhibitor Rgt-018
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-08-14 Classification: SIGNALING PROTEIN Ligands: A1LVJ, EDO |
 |
Crystal Structure Of Nur77 Lbd In Complex With N-(2'-(4-Hydroxypiperidin-1-Yl)-[4,4'-Bipyridin]-2-Yl)Cinnamamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2024-07-24 Classification: TRANSCRIPTION Ligands: A1D59 |
 |
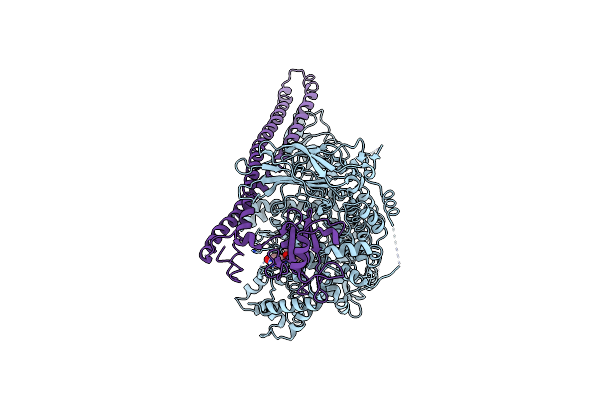
Cryoem Structure Of Human Pi3K-Alpha (P85/P110-H1047R) With Qr-7909 Binding At An Allosteric Site
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: ONCOPROTEIN Ligands: UEX |
 |
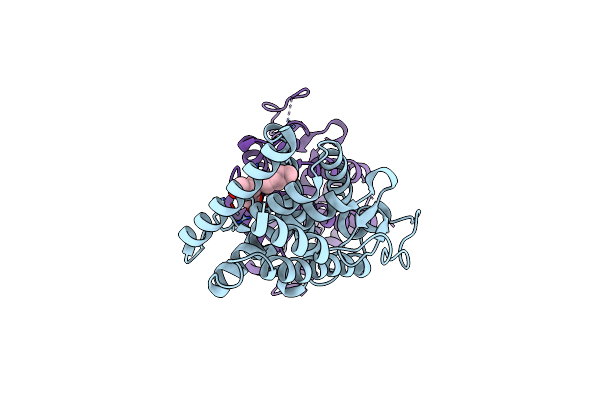
Cryoem Structure Of Human Pi3K-Alpha (P85/P110-H1047R) With Qr-8557 Binding At An Allosteric Site
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: ONCOPROTEIN Ligands: UJ3 |
 |
Crystal Structure Of Tr3 Lbd In Complex With Para-Positioned 3,4,5-Trisubstituted Benzene Derivatives
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-01-24 Classification: TRANSCRIPTION Ligands: XBF |
 |
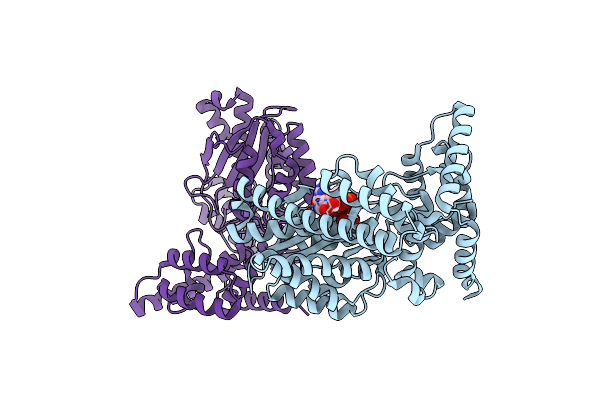
Organism: Legionella
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2023-11-08 Classification: TRANSFERASE |
 |
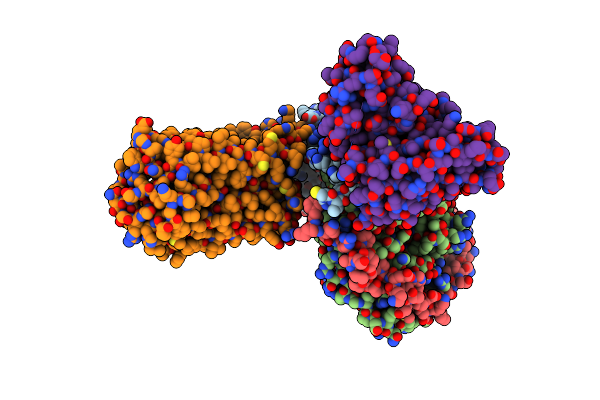
Organism: Legionella pneumophila subsp. pneumophila str. philadelphia 1
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2023-05-17 Classification: TOXIN Ligands: GNP |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: SIGNALING PROTEIN Ligands: IUD |
 |
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: SIGNALING PROTEIN Ligands: CLR |
 |
Organism: Legionella pneumophila subsp. pneumophila str. philadelphia 1
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2023-04-12 Classification: TRANSFERASE |
 |
Organism: Legionella pneumophila subsp. pneumophila str. philadelphia 1
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-04-12 Classification: TRANSFERASE Ligands: UPG |
 |
Complex Structure Of Cdk2/Cyclin E1 And A Potent, Selective Macrocyclic Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2023-02-22 Classification: CELL CYCLE Ligands: WZU |
 |
Complex Structure Of Cdk2/Cyclin E1 And A Potent, Selective Small Molecule Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-02-22 Classification: CELL CYCLE Ligands: WZZ |

