Search Count: 31
 |
The Mutant Variant Of Pngm-1, H93 Was Substituuted For Alanine To Study Metal Coordination
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2023-01-18 Classification: HYDROLASE Ligands: ZN |
 |
The Mutant Variant Of Pngm-1. H279A Was Substituted For Alanine To Study Metal Coordination.
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2021-04-28 Classification: ANTIBIOTIC Ligands: ZN |
 |
The Mutant Variant Of Pngm-1. H96 Was Substituted For Alanine To Study Metal Coordination.
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-04-28 Classification: ANTIBIOTIC Ligands: ZN |
 |
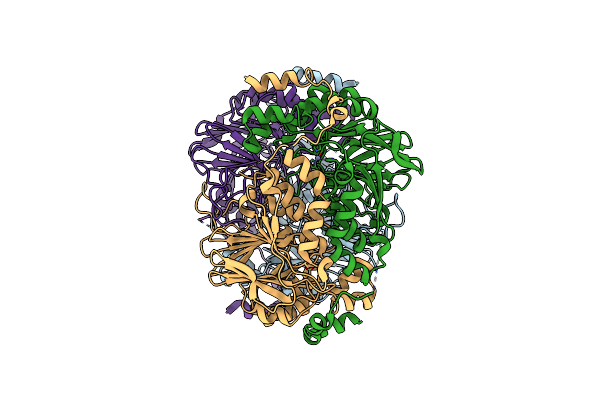
The Mutant Variant Of Pngm-1. H257 Was Substituted For Alanine To Study Substrate Binding.
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-04-28 Classification: ANTIBIOTIC Ligands: ZN |
 |
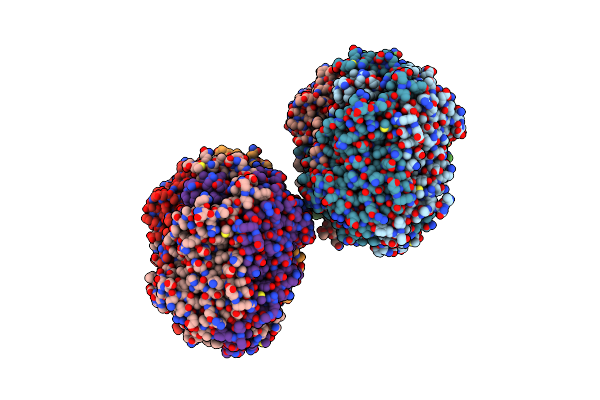
The Mutant Variant Of Pngm-1. H279 Was Substituted For Alanine To Study Metal Coordination.
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2021-04-28 Classification: ANTIBIOTIC Ligands: ZN |
 |
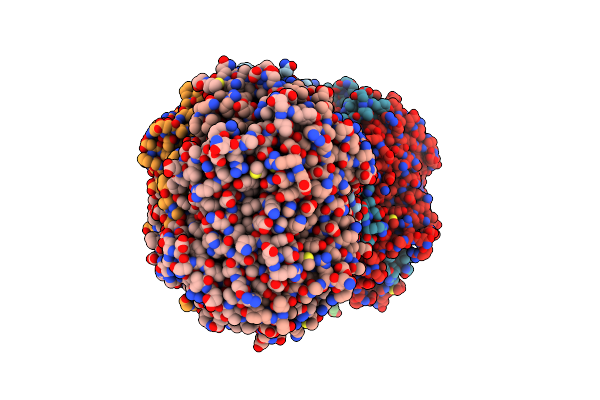
The Mutant Variant Of Pngm-1. H91 Was Substituted For Alanine To Study Metal Coordination.
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-04-28 Classification: ANTIBIOTIC Ligands: ZN |
 |
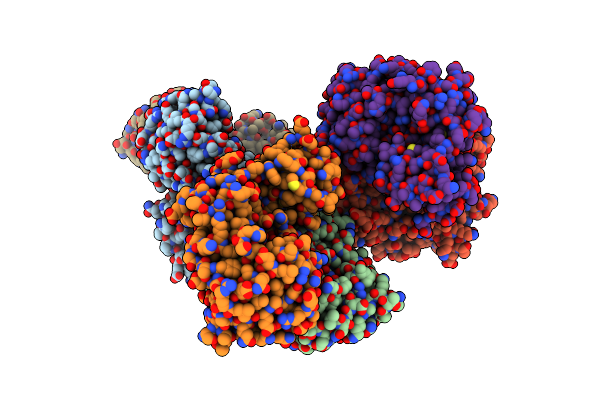
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-02-27 Classification: HYDROLASE Ligands: ZN |
 |
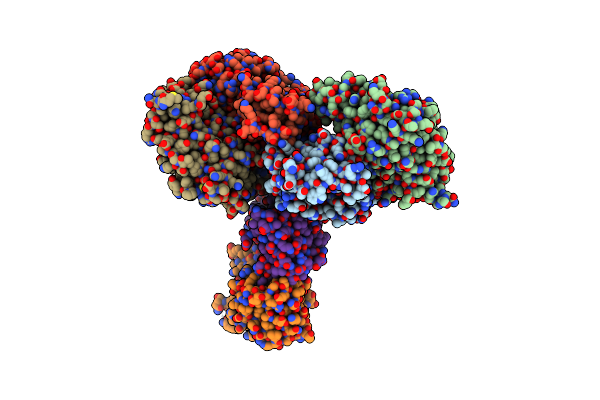
Crystal Structure Of D-Alanine-D-Alanine Ligase From Acinetobacter Baumannii
Organism: Acinetobacter baumannii acicu
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2016-08-17 Classification: LIGASE |
 |
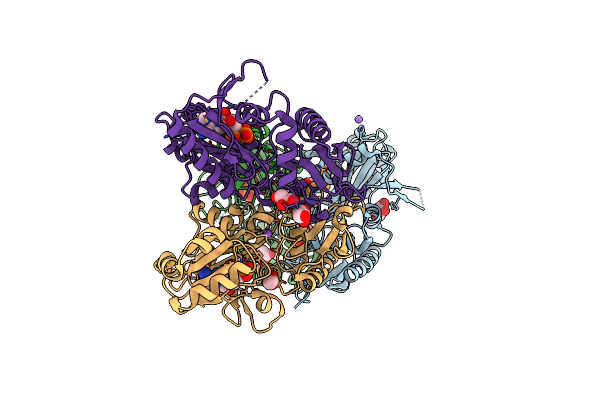
Crystal Structure Of D-Alanine-D-Alanine Ligase From Acinetobacter Baumannii, Space Group P212121
Organism: Acinetobacter baumannii acicu
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2016-08-17 Classification: LIGASE |
 |
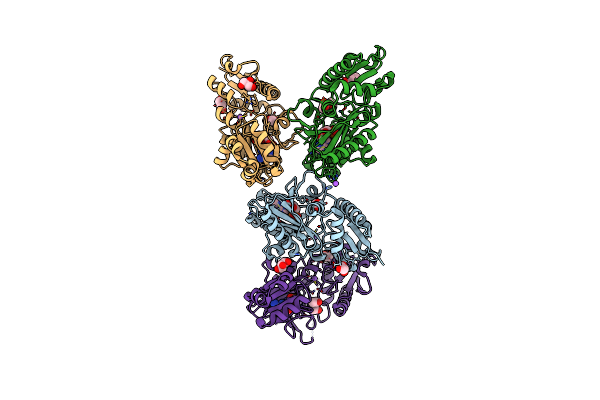
Crystal Structure Of Adp Complexed D-Alanine-D-Alanine Ligase(Ddl) From Yersinia Pestis
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2016-03-02 Classification: LIGASE |
 |
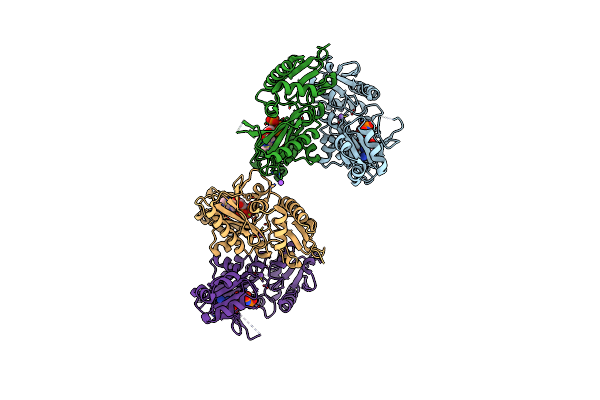
Crystal Structure Of Amp Complexed D-Alanine-D-Alanine Ligase(Ddl) From Yersinia Pestis
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-03-02 Classification: LIGASE Ligands: AMP, NA, ACT, GOL |
 |
Crystal Structure Of Amp-Pnp Complexed D-Alanine-D-Alanine Ligase(Ddl) From Yersinia Pestis
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-03-02 Classification: LIGASE Ligands: ANP, NA, MG |
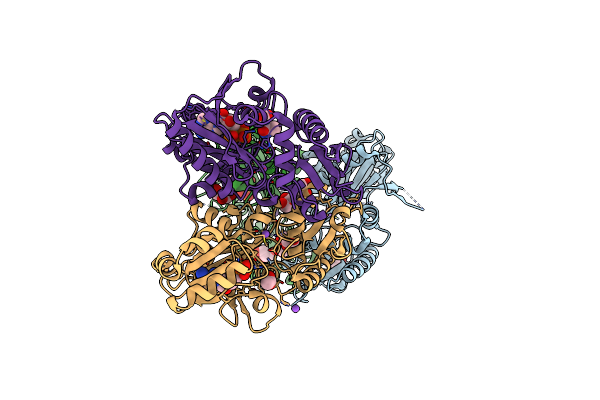 |
Crystal Structure Of Adp And D-Alanyl-D-Alanine Complexed D-Alanine-D-Alanine Ligase(Ddl) From Yersinia Pestis
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-03-02 Classification: LIGASE |
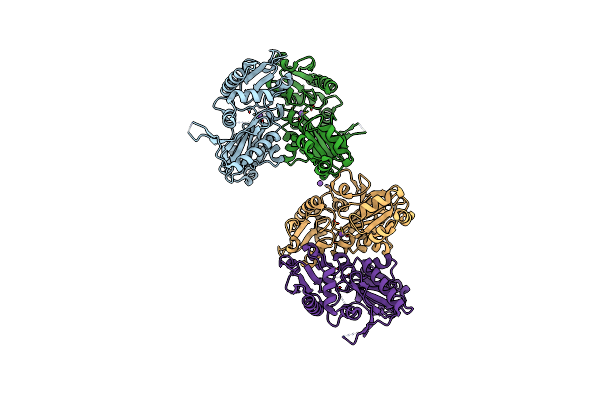 |
Crystal Structure Of Apo D-Alanine-D-Alanine Ligase(Ddl) From Yersinia Pestis
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-01-13 Classification: LIGASE |
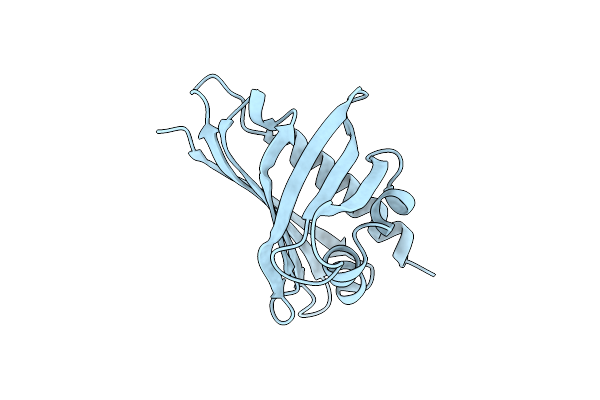 |
Crystal Structure Of Ginseng Major Latex-Like Protein 151 (Glp) From Panax Ginseng. (Crystal-1)
Organism: Panax ginseng
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-05-13 Classification: PROTEIN BINDING |
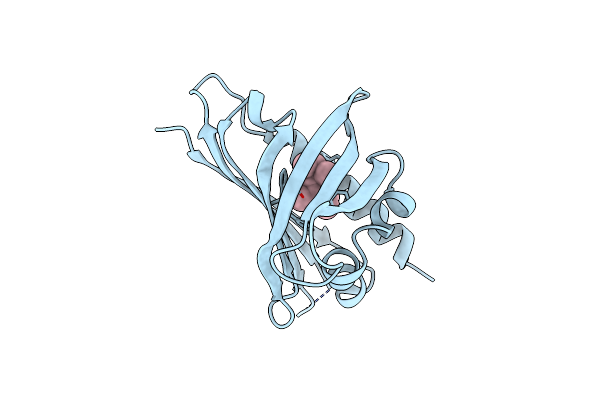 |
Crystal Structure Of Ginseng Major Latex-Like Protein 151 (Glp) From Panax Ginseng. (Crystal-2)
Organism: Panax ginseng
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2015-05-13 Classification: PROTEIN BINDING Ligands: 3MV |
 |
Crystal Structure Of Ginseng Major Latex-Like Protein 151 (Glp) From Panax Ginseng. (Crystal-3)
Organism: Panax ginseng
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2015-05-13 Classification: PROTEIN BINDING |
 |
Structure Of Adc-68, A Novel Carbapenem-Hydrolyzing Class C Extended-Spectrum -Lactamase From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-01-21 Classification: HYDROLASE Ligands: MES, CIT |
 |
Structure And Atypical Hydrolysis Mechanism Of The Nudix Hydrolase Orf153 (Ymfb) From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-05-14 Classification: HYDROLASE Ligands: MN, SO4 |
 |
Structure And Atypical Hydrolysis Mechanism Of The Nudix Hydrolase Orf153 (Ymfb) From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2014-05-14 Classification: HYDROLASE |

