Search Count: 115
 |
Cryo-Em Structure Of Slc30A10 In Mn2+-Bound State, Determined In Inward-Facing Conformation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: TRANSPORT PROTEIN Ligands: MN |
 |
Cryo-Em Structure Of Slc30A10, Determined In Asymmetric Conformations-One Subunit In An Inward-Facing Mn2+-Bound And The Other In An Outward-Facing Mn2+-Unbound Conformation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: TRANSPORT PROTEIN Ligands: MN |
 |
Cryo-Em Structure Of Slc30A10 In The Absence Of Mn2+, Determined In Inward-Facing Conformation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: TRANSPORT PROTEIN |
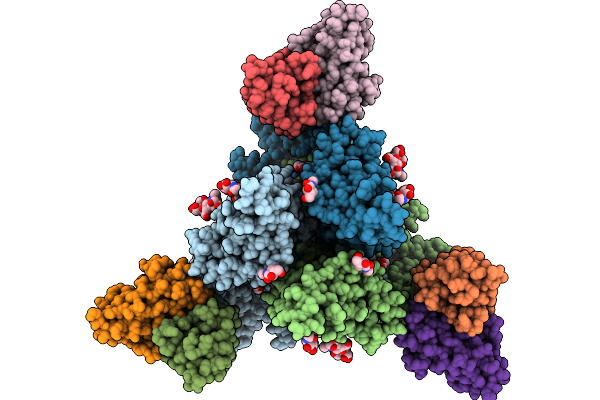 |
Cryo-Em Structure Of Neutralizing Murine Antibody Ws.Hsv-1.24 In Complex With Hsv-1 Glycoprotein B Trimer Gb-Ecto.516P.531E
Organism: Human alphaherpesvirus 1, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: VIRAL PROTEIN Ligands: NAG |
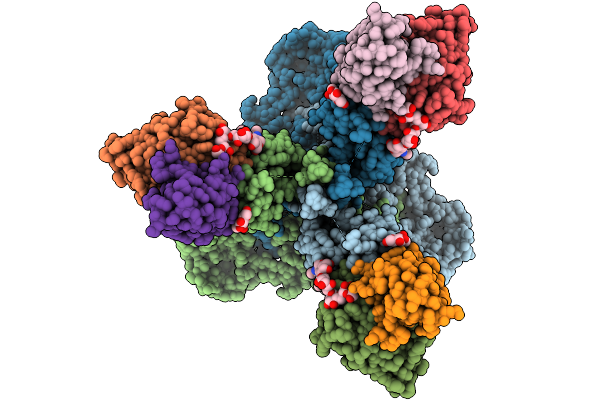 |
Cryo-Em Structure Of Neutralizing Human Antibody D48 In Complex With Hsv-1 Glycoprotein B Trimer Gb-Ecto.516P.531E.Ds
Organism: Human alphaherpesvirus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: VIRAL PROTEIN Ligands: NAG |
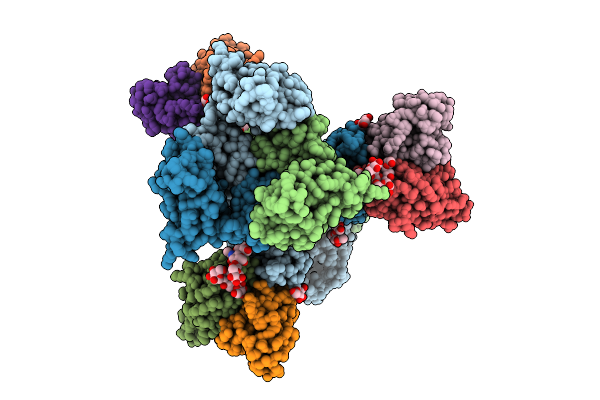 |
Cryo-Em Structure Of Neutralizing Human Antibody D48 In Complex With Hsv-1 Glycoprotein B Trimer Gb-Ecto.516P
Organism: Human alphaherpesvirus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: VIRAL PROTEIN Ligands: NAG |
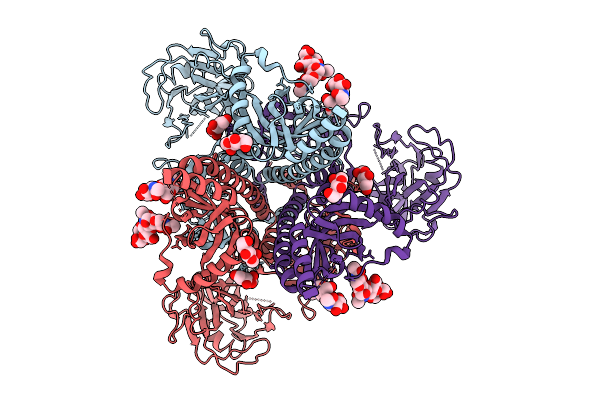 |
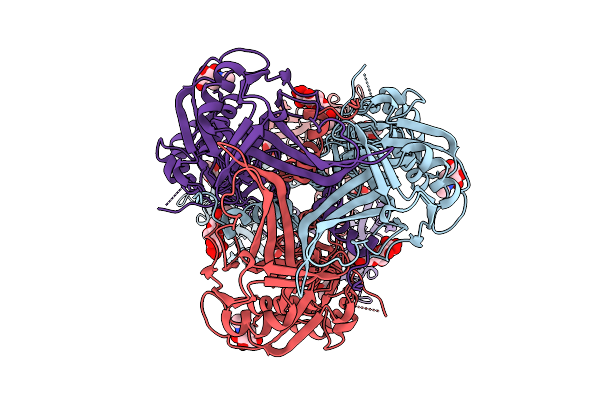
Cryo-Em Structure Of Gb-Ecto.516P.531E.Ds, A Prefusion-Stabilized Hsv-1 Glycoprotein B Extracellular Domain
Organism: Human alphaherpesvirus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Gb-Ecto.516P, An Hsv-1 Glycoprotein B Extracellular Domain
Organism: Human alphaherpesvirus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Local Refinement Of Antibody 19-77 In Complex With Prefusion Sars-Cov-2 Spike Glycoprotein Rbd
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
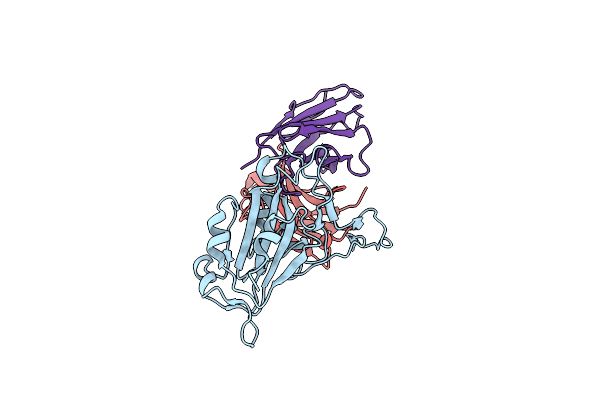 |
Cryo-Em Refinement Of Antibody 19-77 R71V In Complex With Sars-Cov-2 Hk.3 Rbd
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Refinement Of Antibody 19-77 R71V In Complex With Sars-Cov-2 Jd.1.1 Rbd
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
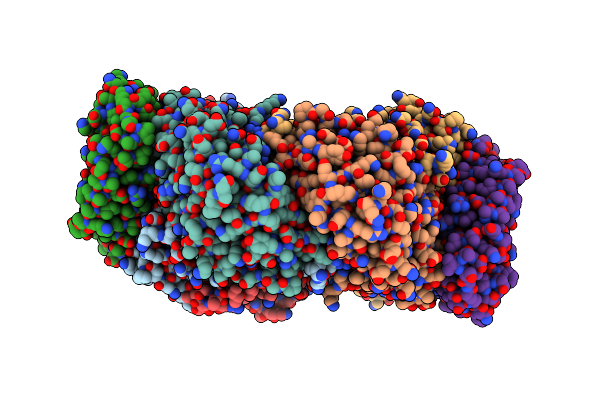 |
Cryo-Em Complex Structure Between Hydroxylase And Regulatory Component From Soluble Methane Monooxygenase
Organism: Methylosinus sporium
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: OXIDOREDUCTASE Ligands: FE |
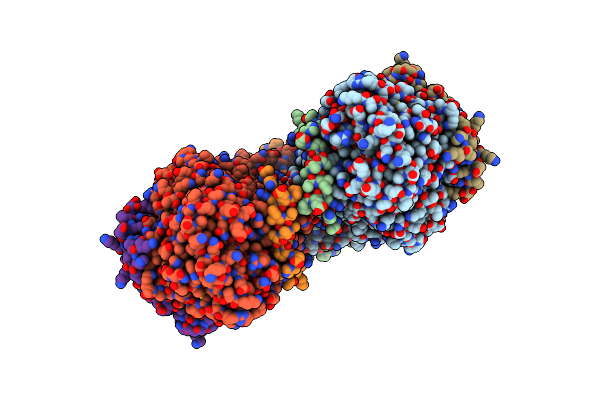 |
Cryo-Em Structure Of Hydroxylase In Soluble Methane Monooxygenase From Methylosinus Sporium 5
Organism: Methylosinus sporium
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2025-04-23 Classification: HYDROLASE |
 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: GDP, ALF, MG |
 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: GDP, MG |
 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: GDP, LMR |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2025-03-19 Classification: PROTEIN BINDING |
 |
Organism: Micromonospora pattaloongensis
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: EDO, GOL |

