Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
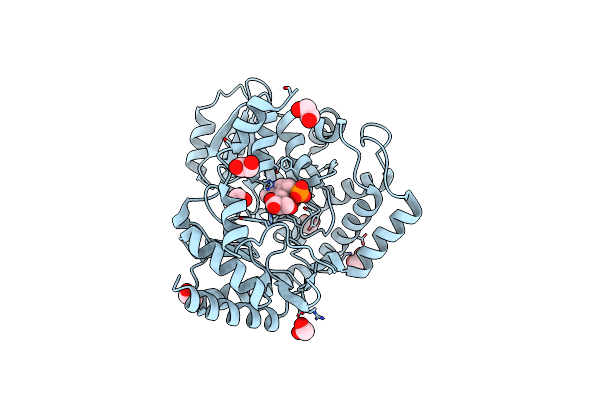 |
Crystal Structure Of Serine Palmitoyltransferase Soaked In 190 Mm D-Serine Solution
Organism: Sphingobacterium multivorum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-04-10 Classification: TRANSFERASE Ligands: PLS, EDO |
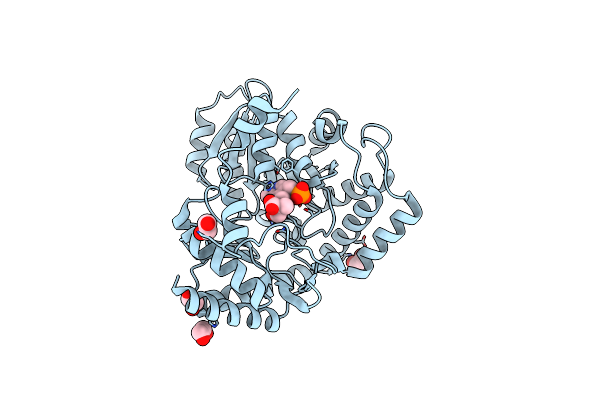 |
Crystal Structure Of Serine Palmitoyltransferase Complexed With D-Methylserine
Organism: Sphingobacterium multivorum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-04-10 Classification: TRANSFERASE Ligands: S5R, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-08-26 Classification: METAL BINDING PROTEIN Ligands: EF2, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-08-26 Classification: METAL BINDING PROTEIN Ligands: F4U, ZN, SO4 |
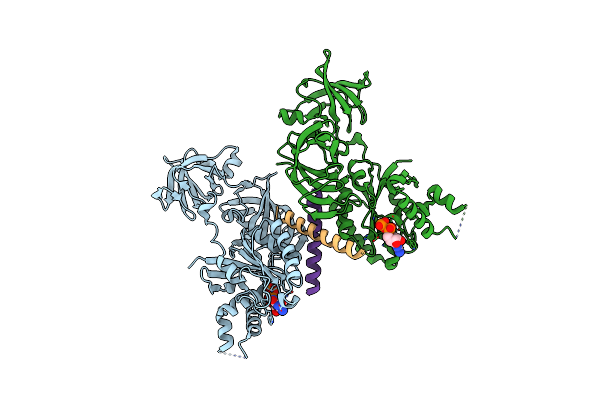 |
Crystal Structure Of A Complex Of The Archaeal Ribosomal Stalk Protein Ap1 And The Gdp-Bound Archaeal Elongation Factor Aef1Alpha
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-12-24 Classification: TRANSLATION/RIBOSOMAL PROTEIN Ligands: GDP |
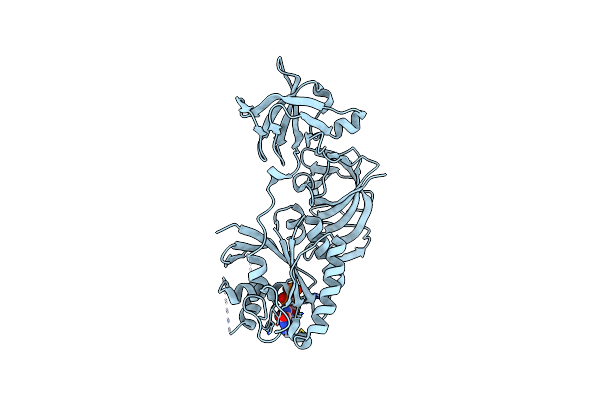 |
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2014-12-24 Classification: TRANSLATION Ligands: GDP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:0.90 Å Release Date: 2012-09-26 Classification: CELL ADHESION |
 |
Crystal Structure Of The Ppargamma-Lbd Complexed With A Cercosporamide Derivative Modulator
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-08-08 Classification: TRANSCRIPTION/TRANSCRIPTION REGULATOR Ligands: FCM |
 |
Crystal Structure Of The Ppargamma-Lbd Complexed With A Cercosporamide Derivative Modulator
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2012-02-01 Classification: TRANSCRIPTION/TRANSCRIPTION REGULATOR Ligands: 17L |
 |
Crystal Structure Of The Ppargamma-Lbd Complexed With A Cercosporamide Derivative Modulator
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-02-01 Classification: TRANSCRIPTION/TRANSCRIPTION REGULATOR Ligands: 21L |
 |
Crystal Structure Of The Ppargamma-Lbd Complexed With A Cercosporamide Derivative Modulator
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-02-01 Classification: TRANSCRIPTION/TRANSCRIPTION REGULATOR Ligands: 24L |
 |
Organism: Vibrio parahaemolyticus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-03-31 Classification: TOXIN |
 |
Crystal Structure Of Protein Ph1601P In Complex With Protein Ph1771P Of Archaeal Ribonuclease P From Pyrococcus Horikoshii Ot3
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2008-10-14 Classification: HYDROLASE Ligands: NO3, GOL, ZN |
 |
Crystal Structure Analysis Of Protein Component Ph1496P Of P.Horikoshii Ribonuclease P
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-04-25 Classification: RIBOSOME, HYDROLASE |
 |
Crystal Structure Of Ribosome Recycling Factor From Vibrio Parahaemolyticus
Organism: Vibrio parahaemolyticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2003-06-17 Classification: TRANSLATION |