Search Count: 30
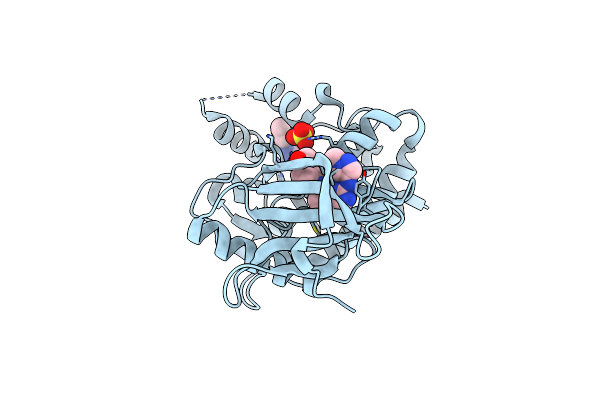 |
Human Jak3 Kinase Domain With 1-(4-((2-((1-Methyl-1H-Pyrazol-4-Yl)Amino)Quinazolin-8-Yl)Amino)Piperidin-1-Yl)Ethan-1-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-02-02 Classification: SIGNALING PROTEIN Ligands: 934, SO4, PHU |
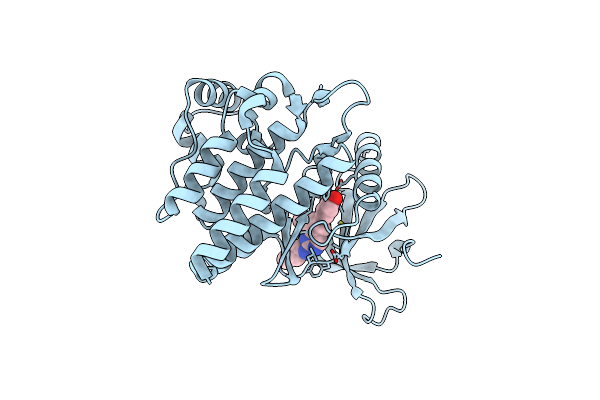 |
Jak2 In Complex With 4-{8-Methoxy-2-[(1-Methyl-1H-Pyrazol-4-Yl)Amino]Quinazolin-6-Yl}Phenol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2022-02-02 Classification: SIGNALING PROTEIN Ligands: 9I8 |
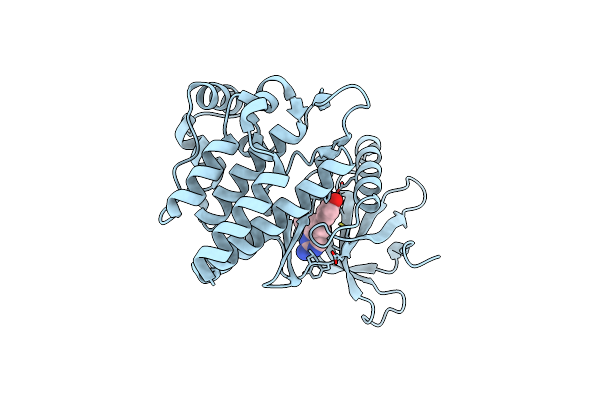 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2022-02-02 Classification: SIGNALING PROTEIN Ligands: 9I5 |
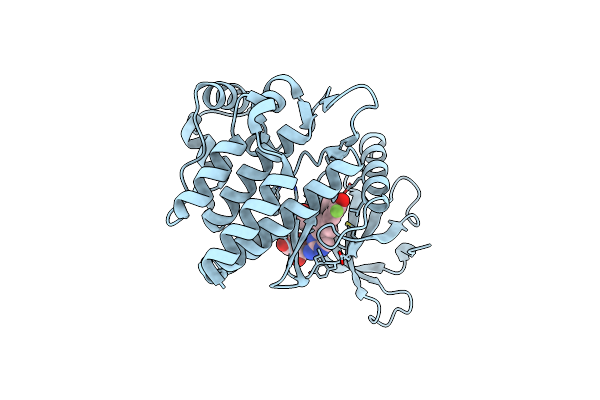 |
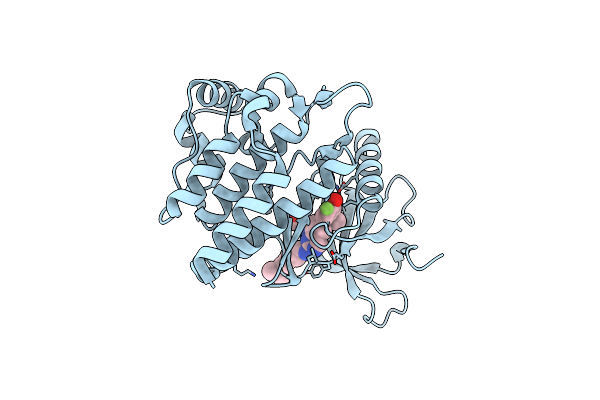
Jak2 In Complex With 4-(2-Amino-8-{[(2S)-1-Hydroxypropan-2-Yl]Amino}Quinazolin-6-Yl)-5-Ethyl-2-Fluorophenol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2022-02-02 Classification: SIGNALING PROTEIN Ligands: 9I2, GOL |
 |
Jak2 In Complex With 4-(2-{[5-(Dimethylamino)Pentyl]Amino}-8-{[(2S)-1-Hydroxypropan-2-Yl]Amino}Quinazolin-6-Yl)-5-Ethyl-2-Fluorophenol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-02-02 Classification: SIGNALING PROTEIN Ligands: 9HR |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, CL, GOL |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-(5-Chloro-6-Methyl-2-Oxo-2,3-Dihydro-1,3-Benzoxazol-3-Yl)Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, CL, 8RK, GOL |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-{5-Chloro-2-Oxo-6-[(1R)-1-(Pyridin-2-Yl)Ethoxy]-2,3-Dihydro-1,3-Benzoxazol-3-Yl}Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, 8R8 |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-{5-Chloro-6-[(1R)-1-(Pyridin-2-Yl)Ethoxy]-1,2-Benzoxazol-3-Yl}Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, 8R5 |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-{5-Chloro-6-[(1R)-1-(6-Methylpyridazin-3-Yl)Ethoxy]-1,2-Benzoxazol-3-Yl}Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, 8RB |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With The Enzyme Substrate L-Kynurenine
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, CL, GOL, KYN |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2016-09-21 Classification: TRANSFERASE |
 |
Organism: Staphylococcus aureus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-12-16 Classification: ISOMERASE Ligands: MN, GOL, SO4, NA, 54Q, DT |
 |
Organism: Staphylococcus aureus, Staphylococcus aureus (strain n315), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2015-12-16 Classification: ISOMERASE Ligands: SO4, MN, GOL, NA, EVP |
 |
Organism: Staphylococcus aureus (strain n315), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2015-12-16 Classification: ISOMERASE Ligands: SO4, NA, GOL, MN, 53M, 53L, 50M |
 |
Organism: Staphylococcus aureus (strain n315), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2015-12-16 Classification: ISOMERASE |
 |
Organism: Staphylococcus aureus (strain n315), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2015-12-16 Classification: HYDROLASE Ligands: MG, GOL, MFX |
 |
Organism: Staphylococcus aureus (strain n315), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2015-12-16 Classification: ISOMERASE Ligands: MN, GOL |
 |
X-Ray Crystal Structure At 2.20A Resolution Of Human Mitochondrial Branched Chain Aminotransferase (Bcatm) Complexed With A Pyrazolopyrimidinone Fragment And An Internal Aldimine Linked Plp.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-07-01 Classification: TRANSFERASE Ligands: PLP, 4VT, CL, EDO |
 |
X-Ray Crystal Structure At 2.20A Resolution Of Human Mitochondrial Branched Chain Aminotransferase (Bcatm) Complexed With A Pyrazolopyrimidinone Fragment And An Internal Aldimine Linked Plp.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-07-01 Classification: TRANSFERASE Ligands: PLP, 4VS, CL, EDO, GOL |

