Search Count: 7
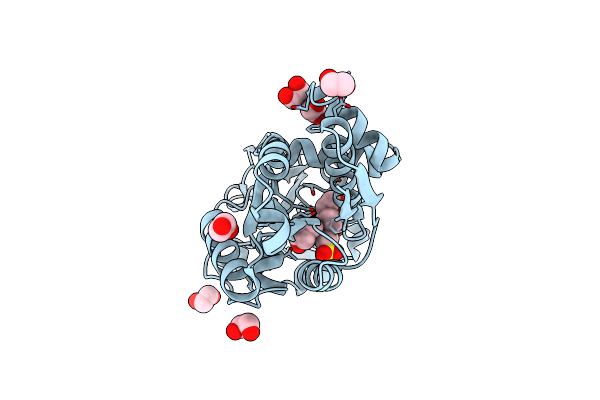 |
A Molecular Switch Changes The Low To The High Affinity State In The Substrate Binding Protein Afprox
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-04-06 Classification: TRANSPORT PROTEIN Ligands: BET, EDO, EPE |
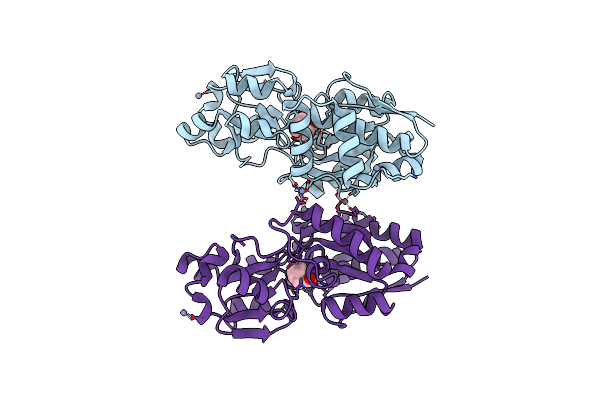 |
Crystal Structure Of Prox From Archeoglobus Fulgidus In Complex With Proline Betaine
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-09-14 Classification: PROTEIN BINDING Ligands: ZN, PBE |
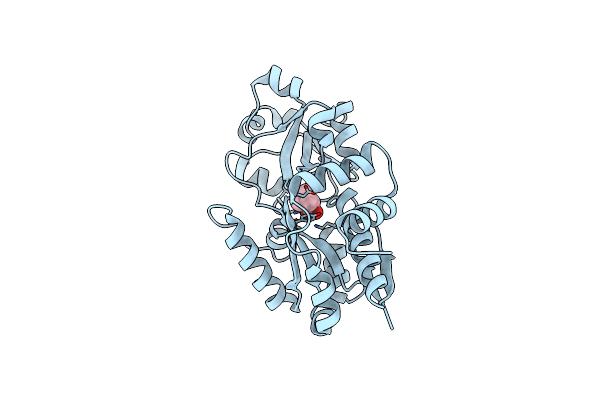 |
Crystal Structure Of Prox From Archeoglobus Fulgidus In Complex With Glycine Betaine
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-09-14 Classification: PROTEIN BINDING Ligands: BET |
 |
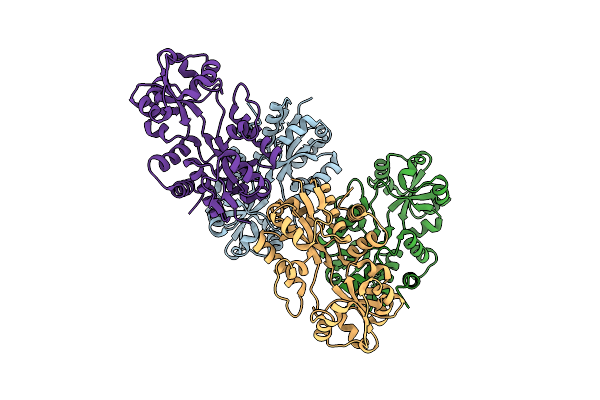
Crystal Structure Of Prox From Archeoglobus Fulgidus In Complex With Trimethyl Ammonium
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-09-14 Classification: PROTEIN BINDING Ligands: ZN, CL, TMA |
 |
Crystal Structure Of Prox From Archeoglobus Fulgidus In The Ligand Free Form
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-09-14 Classification: PROTEIN BINDING Ligands: CL, MG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2004-02-24 Classification: PROTEIN BINDING Ligands: UNX, BET |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2004-02-24 Classification: PROTEIN BINDING Ligands: UNX, PBE |

