Search Count: 118
 |
Organism: Bacillus paranthracis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PROTEIN FIBRIL |
 |
Organism: Bacillota bacterium
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: PROTEIN FIBRIL |
 |
Organism: Pseudomonas sp. uk4
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: PROTEIN FIBRIL |
 |
Organism: Pseudomonas fluorescens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: STRUCTURAL PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2025-05-28 Classification: RIBOSOME Ligands: OHX, MG, ZN, K, ZWB, SPD |
 |
C-Terminal Domain Of The F-Ena Tip Fibrillum F-Bcla From Bacillus Thuringiensis
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2025-03-12 Classification: STRUCTURAL PROTEIN Ligands: GOL, EDO |
 |
F-Ena Exosporium Anchoring Complex Between Exsf And A Peptide Derived From The N-Terminus Of F-Anchor
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2025-03-12 Classification: STRUCTURAL PROTEIN |
 |
Cryoem Structure Of F-Ena Fibers On The Spores Of Bacillus Thuringiensis Serovar Kurstaki
Organism: Bacillus thuringiensis serovar kurstaki
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: PROTEIN FIBRIL |
 |
Organism: Bacillus thuringiensis serovar kurstaki
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: PROTEIN FIBRIL |
 |
Organism: Cohnella sp. ov330
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: PROTEIN FIBRIL |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-11-27 Classification: RIBOSOME Ligands: OHX, MG, SPD, XBI, ZN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.16 Å Release Date: 2024-10-16 Classification: RIBOSOME Ligands: OHX, MG, K, SPD, X1K, ZN, OS |
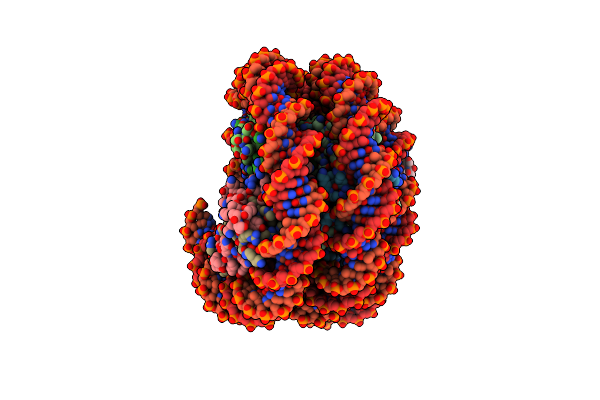 |
Structure Of A Hexasome-Nucleosome Complex With A Dyad-To-Dyad Distance Of 103 Bp.
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: DNA BINDING PROTEIN |
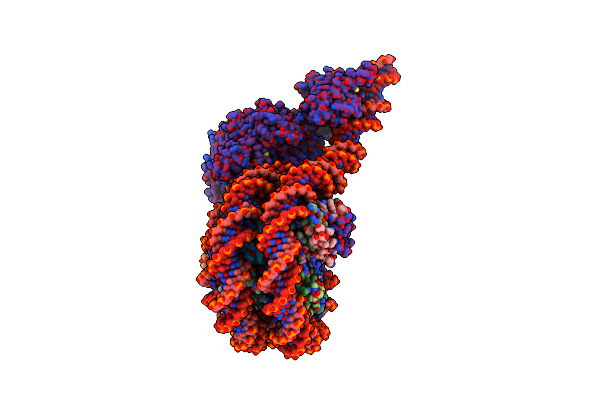 |
Structure Of Chd1 Bound To A Hexasome-Nucleosome Complex With A Dyad-To-Dyad Distance Of 103 Bp.
Organism: Xenopus laevis, Synthetic construct, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: DNA BINDING PROTEIN Ligands: ADP, BEF, MG |
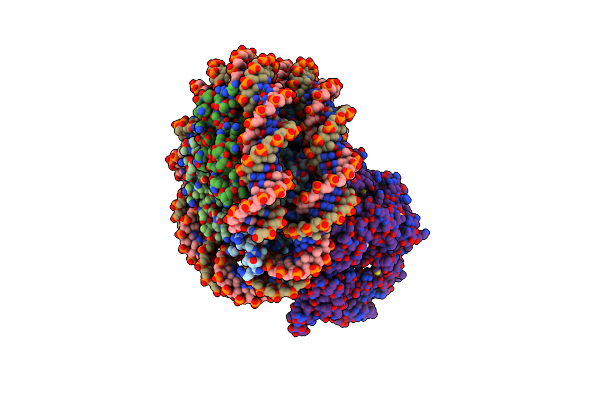 |
Structure Of Chd1 Bound To A Dinucleosome With A Dyad-To-Dyad Distance Of 103 Bp.
Organism: Xenopus laevis, Synthetic construct, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: DNA BINDING PROTEIN Ligands: ADP, BEF, MG |
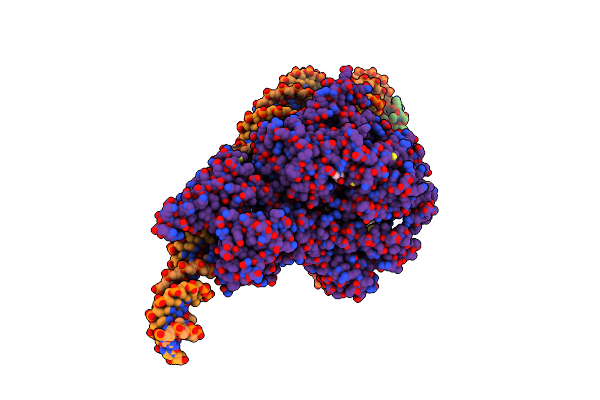 |
Structure Of A Mononucleosome Bound By One Copy Of Chd1 With The Dbd On The Exit-Side Dna.
Organism: Xenopus laevis, Synthetic construct, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: DNA BINDING PROTEIN Ligands: ADP, BEF, MG |
 |
Organism: Candidatus nitrospira inopinata
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2024-08-14 Classification: METAL BINDING PROTEIN Ligands: SO4, MN, NI |
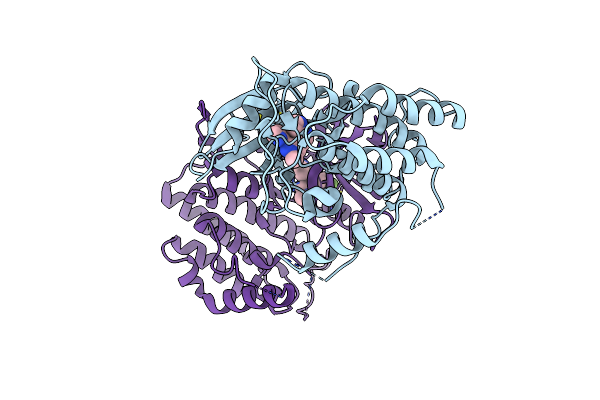 |
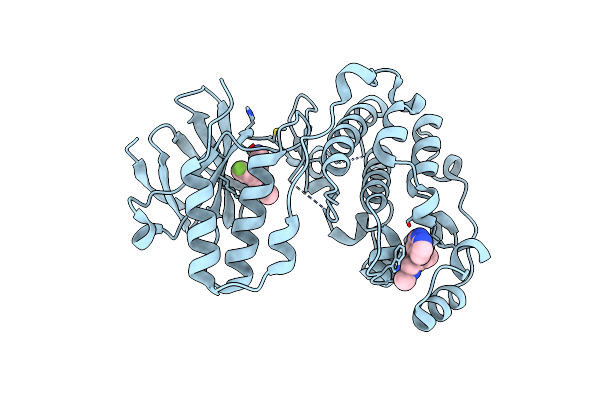
Structure Of Casein Kinase I Isoform Delta (Ck1D) Complexed With Inhibitor 7
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2024-05-08 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: A1AD7 |
 |
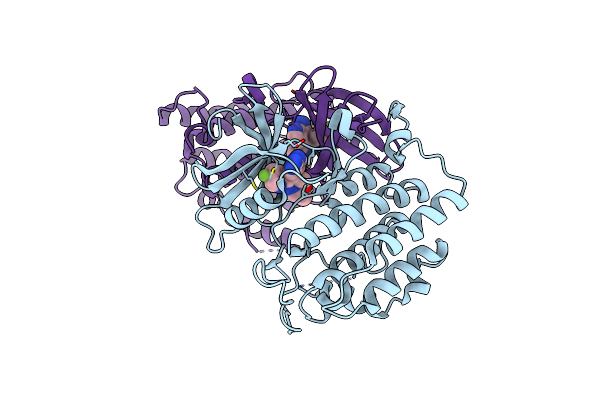
Structure Of P38 Alpha (Mitogen-Activated Protein Kinase 14) Complexed With Inhibitor 6
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-05-08 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: A1AD9 |
 |
Structure Of Casein Kinase I Isoform Delta (Ck1D) Complexed With Inhibitor 15
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2024-05-08 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: A1AD8 |

