Search Count: 3
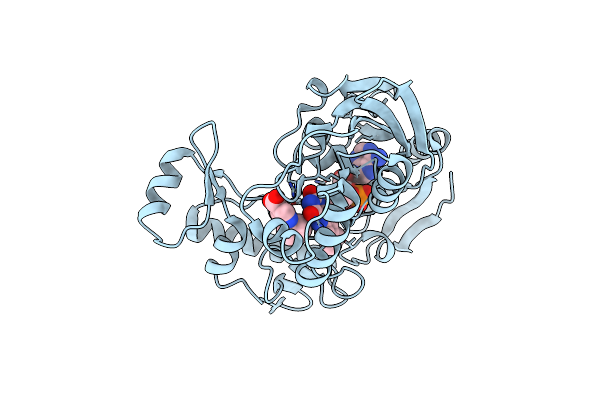 |
Crystal Structure Of Udp-N-Acetylenolpyruvoylglucosamine Reductase (Murb) From Pseudomonas Aeruginosa In Complex With Fad And A Pyrazole Derivative (Fragment 4)
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2021-11-03 Classification: OXIDOREDUCTASE Ligands: FAD, 9FH |
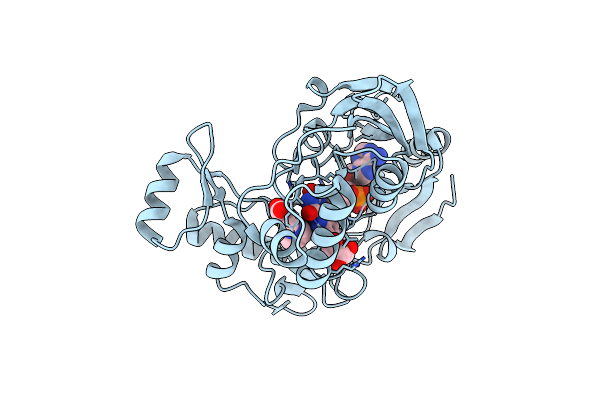 |
Crystal Structure Of Udp-N-Acetylenolpyruvoylglucosamine Reductase (Murb) From Pseudomonas Aeruginosa In Complex With Fad And A Pyrazole Derivative (Fragment 18)
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-11-03 Classification: OXIDOREDUCTASE Ligands: 0IM, GOL, FAD |
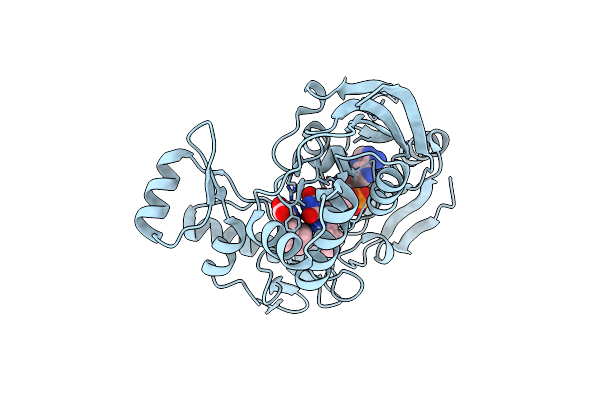 |
Crystal Structure Of Udp-N-Acetylenolpyruvoylglucosamine Reductase (Murb) From Pseudomonas Aeruginosa In Complex With Fad And A Pyrazole Derivative (Fragment 18)
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2021-11-03 Classification: OXIDOREDUCTASE Ligands: FAD, 0JI |

