Search Count: 13
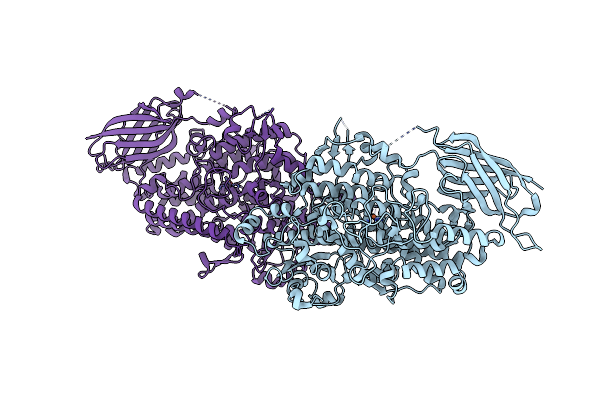 |
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FE |
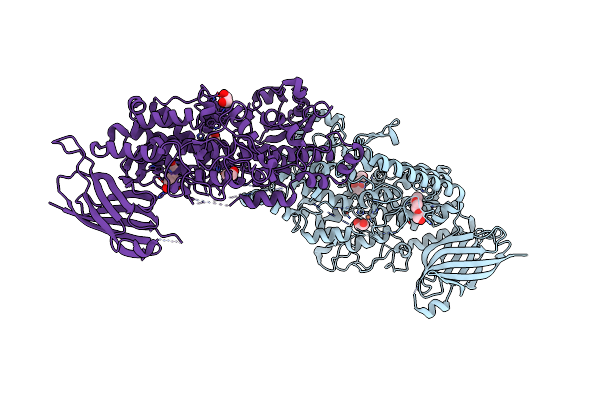 |
Vitis Vinifera Dimeric 13S-Lipoxygenase Loxa In A Detergent Bound Open Conformation
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FE, PQE, EDO |
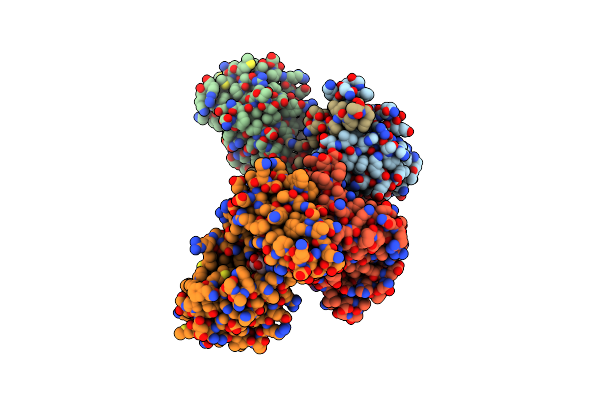 |
Changes In Conformational Equilibria Regulate The Activity Of The Dcp2 Decapping Enzyme
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2017-05-24 Classification: RNA BINDING PROTEIN Ligands: MG, M7G |
 |
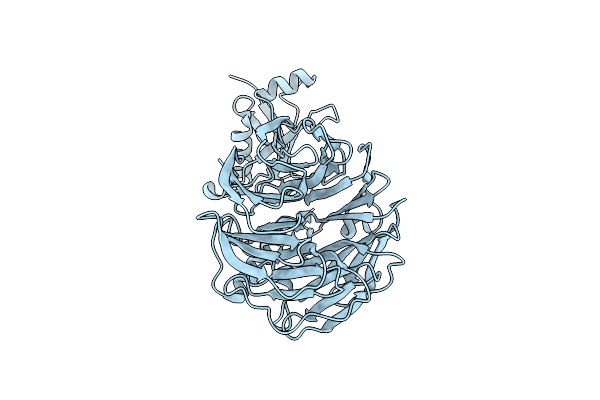
Structural Insight Into An Essential Assembly Factor Network On The Pre-Ribosome
Organism: Chaetomium thermophilum
Method: SOLUTION NMR Release Date: 2014-12-03 Classification: RIBOSOMAL PROTEIN |
 |
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-11-19 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-11-19 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL |
 |
Organism: Saccharomyces cerevisiae, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2014-11-19 Classification: PROTEIN BINDING Ligands: SO4 |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2014-07-09 Classification: RIBOSOME |
 |
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-03-26 Classification: UNKNOWN FUNCTION Ligands: SO4 |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2012-01-11 Classification: TRANSPORT PROTEIN |
 |
Structural Basis For Specific Substrate Recognition By The Chloroplast Signal Recognition Particle Protein Cpsrp43
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2008-08-12 Classification: PROTEIN TRANSPORT, MEMBRANE PROTEIN Ligands: MG |
 |
Structural Basis For Specific Substrate Recognition By The Chloroplast Signal Recognition Particle Protein Cpsrp43
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2008-08-12 Classification: PROTEIN TRANSPORT, MEMBRANE PROTEIN Ligands: CL |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2007-12-25 Classification: PROTEIN TRANSPORT Ligands: MLI |

