Search Count: 397
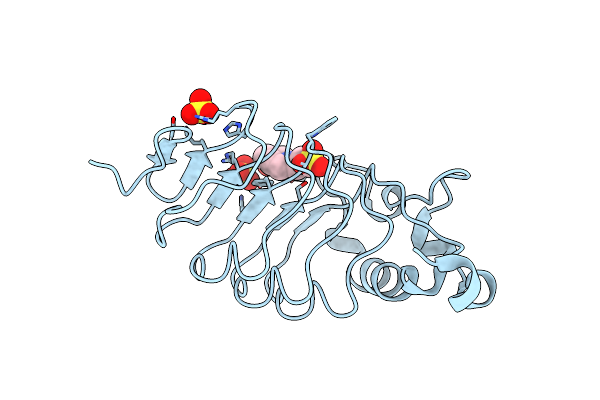 |
The High-Resolution Structure Of A Variable Lymphocyte Receptor From Petromyzon Marinus Capable Of Binding To The Brain Extracellular Matrix
Organism: Petromyzon marinus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2024-07-31 Classification: IMMUNE SYSTEM Ligands: EP1, SO4 |
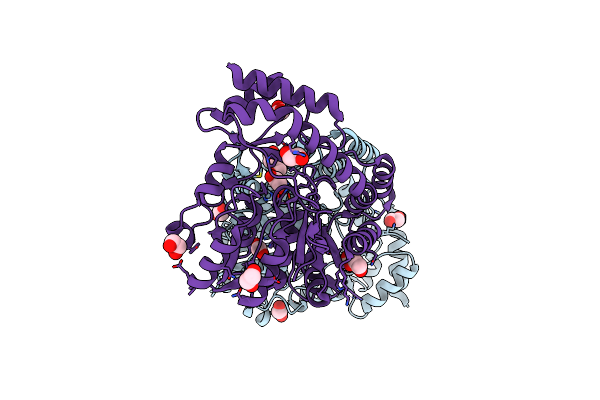 |
X-Ray Structure Of The Aminotransferase From Vibrio Vulnificus Responsible For The Biosynthesis Of 2,3-Diacetamido-4-Amino-2,3,4-Trideoxy-Arabinose In The Presence Of Its Internal Aldimine
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: EDO, CL |
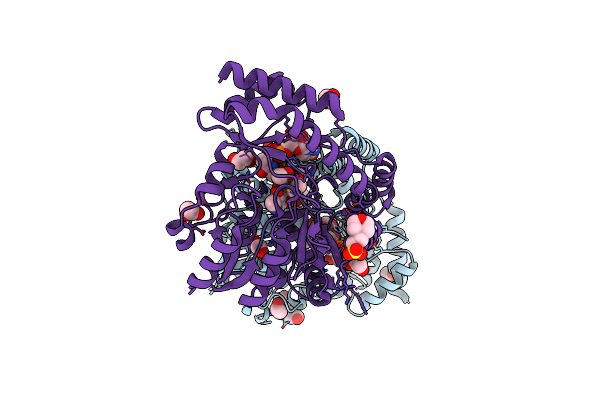 |
X-Ray Structure Of The Aminotransferase From Vibrio Vulnificus Responsible For The Biosynthesis Of 2,3-Diacetamido-4-Amino-2,3,4-Trideoxy-Arabinose In The Presence Of Its External Aldimine With 2,3-Diacetamido-4-Amino-2,3,4-Trideoxy-L-Arabinose
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: A1APE, MES, EDO, PGE |
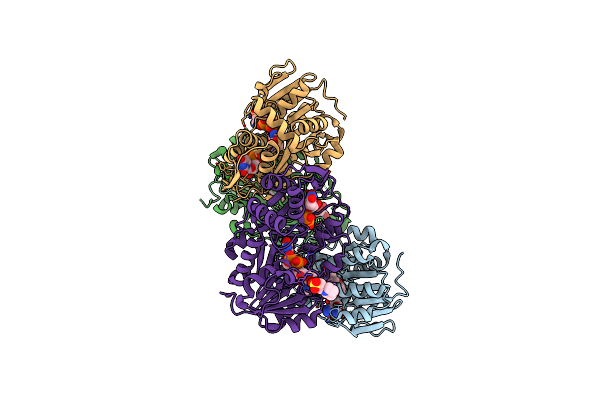 |
Crystal Structure Of The Pcryo_0617 Oxidoreductase/Decarboxylase From Psychrobacter Cryohalolentis K5 In The Presence Of Nad And Udp
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-01-31 Classification: OXIDOREDUCTASE Ligands: NAD, UDP, NA, MES |
 |
Crystal Structure Of The Pcryo_0618 Aminotransferase From Psychrobacter Cryohalolentis K5 In The Presence Of Its Internal Aldimine
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-31 Classification: TRANSFERASE Ligands: EDO, NA, A1ADI, CL |
 |
Crystal Structure Of The Pcryo_0618 Aminotransferase From Psychrobacter Cryohalolentis K5 In The Presence Of Pmp And Glutamate
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-01-31 Classification: TRANSFERASE Ligands: PMP, A1ADI, CL, EDO, MG, PDG, NA |
 |
Crystal Structure Of The Pcryo_0619 N-Acetryltransferase From Psychrobacter Cryohalolentis K5 In The Presence Of Coa-Disulfide
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-01-31 Classification: TRANSFERASE Ligands: 5NG |
 |
Crystal Structure Of The Pcryo_0619 N-Acetyltransferase From Psychrobacter Cryohalolentis K5 Int He Presence Of Acetyl Coenzyme A
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-01-31 Classification: TRANSFERASE Ligands: ACO, COA, EDO, EP1, PAP |
 |
Crystal Structure Of The Pcryo_0619 N-Acetyltransferase From Psychrobacter Cryohalolentis K5
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-01-31 Classification: TRANSFERASE Ligands: SO4, MPD, EDO |
 |
X-Ray Structure Of The Nadp-Dependent Reductase From Campylobacter Jejuni Responsible For The Synthesis Of Cdp-Glucitol In The Presence Of Cdp And Nadp
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-12-20 Classification: OXIDOREDUCTASE Ligands: NAP, CDP, CL, EDO |
 |
X-Ray Structure Of The Nadp-Dependent Reductase From Campylobacter Jejuni Responsible For The Synthesis Of Cdp-Glucitol In The Presence Of Cdp-Glucitol
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-12-20 Classification: OXIDOREDUCTASE Ligands: YCX, CL, PO4, NA |
 |
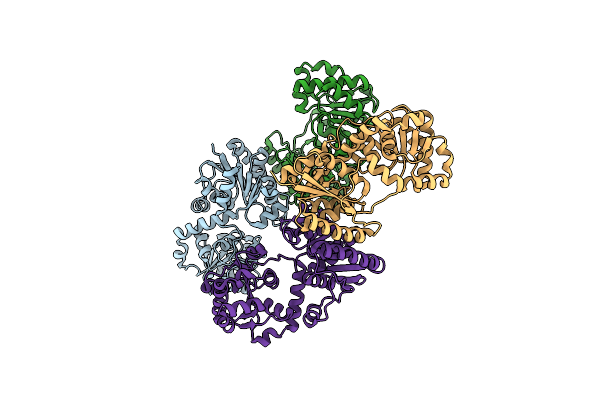
X-Ray Crystal Structure Of Udp- 2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27, Apo Form, Ph 9
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: CL |
 |
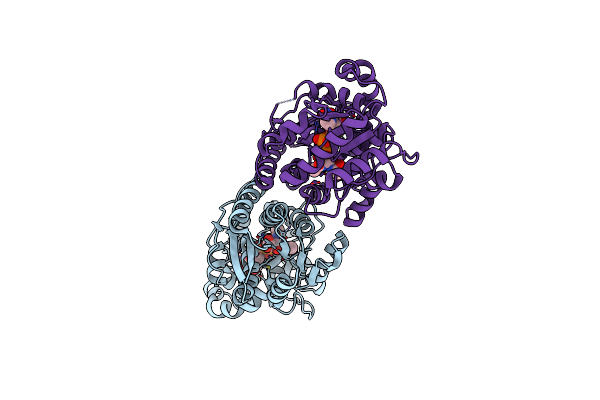
X-Ray Crystal Structure Of Udp- 2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27, D98N Mutation, Apo Structure At Ph 6
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: CL, NA |
 |
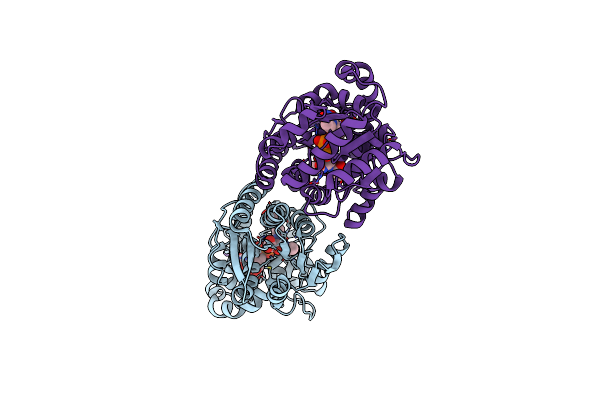
X-Ray Crystal Structure Of Udp- 2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27 In Complex With Its Product Udp-2,3-Diacetamido-2,3-Dideoxy-D-Mannuronic Acid At Ph 5
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: MJ3, CL |
 |
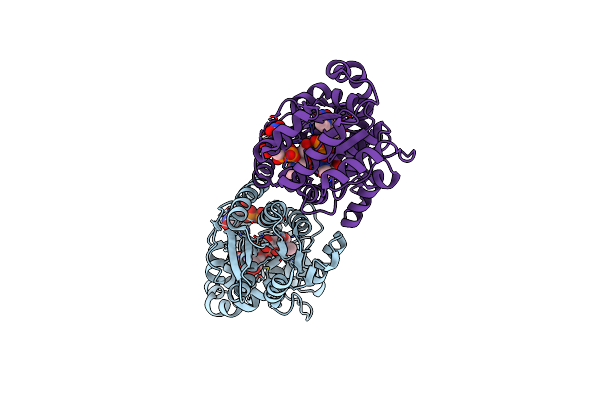
X-Ray Crystal Structure Of Udp- 2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27 In Complex With Its Product Udp-2,3-Diacetamido-2,3-Dideoxy-D-Mannuronic Acid At Ph 9
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: MJ3, EDO, NA, CL |
 |
X-Ray Crystal Structure Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27, D98N Variant In The Presence Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid, Udp-N-Acetylglucosamine And Udp At Ph 7
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: MJL, UDP, UD1 |
 |
X-Ray Crystal Structure Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27, D98N Variant In The Presence Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid And Udp At Ph 9
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: MJL, UDP |
 |
X-Ray Crystal Structure Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27, D98N Variant In The Presence Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid And Udp-N-Acetylglucosamine At Ph 9
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: MJL, UD1, CL, NA |
 |
X-Ray Crystal Structure Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27, D98N Variant In The Presence Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid And Udp-N-Acetylglucosamine At Ph 6
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: MJL, UD1, CL |
 |
X-Ray Crystal Structure Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid-2-Epimerase From Thermus Thermophilus Strain Hb27, D98N Variant In The Presence Of Udp-2,3-Diacetamido-2,3-Dideoxy-Glucuronic Acid And Udp At Ph 6
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-09-13 Classification: ISOMERASE Ligands: MJL, UDP, CL |

