Search Count: 17
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-11-06 Classification: IMMUNE SYSTEM Ligands: SO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: IMMUNE SYSTEM |
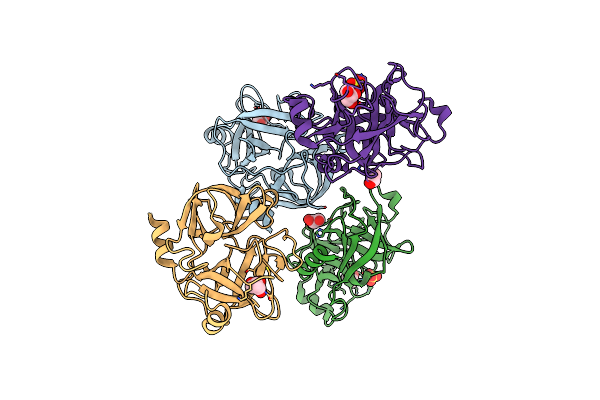 |
Crystal Structure Of The Inhibitor-Free Form Of The Serine Protease Kallikrein-4
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2019-07-17 Classification: HYDROLASE Ligands: GOL, SO4 |
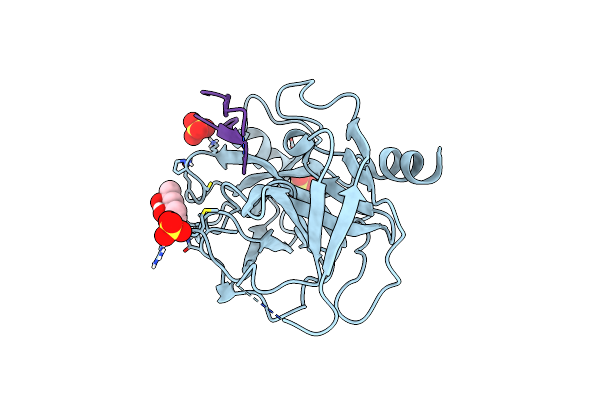 |
Crystal Structure Of Human Klk4 In Complex With Cleaved Sfti-Fcqr(Asn14)[1,14] Inhibitor
Organism: Homo sapiens, Helianthus annuus
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2019-03-13 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: SO4, MPD |
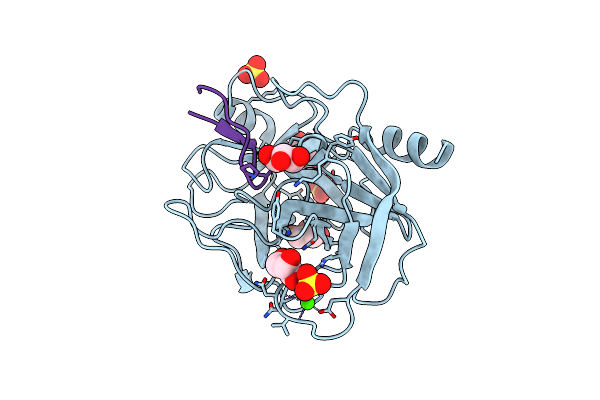 |
Trypsin Complexed With A Modified Sunflower Trypsin Inhibitor, Sfti-Tctr(N12,N14)
Organism: Helianthus annuus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2018-12-19 Classification: HYDROLASE/INHIBITOR Ligands: CA, GOL, SO4 |
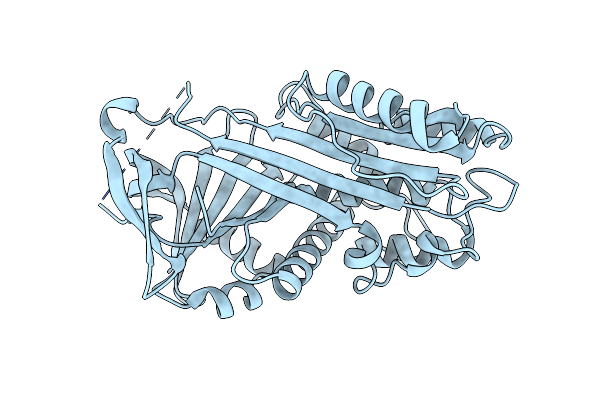 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2018-08-29 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2016-08-17 Classification: protein binding/hydrolase |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2016-08-17 Classification: PROTEIN BINDING Ligands: ZN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-07-20 Classification: hydrolase inhibitor |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2016-07-20 Classification: hydrolase inhibitor Ligands: GOL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-07-20 Classification: hydrolase inhibitor |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2015-02-11 Classification: DE NOVO PROTEIN |
 |
Atomic Resolution Crystal Structure Of Kallikrein-Related Peptidase 4 Complexed With A Modified Sfti Inhibitor Fcqr(N)
Organism: Homo sapiens, Helianthus annuus
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2014-04-30 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2014-04-30 Classification: HYDROLASE Ligands: NI, EDO |
 |
Atomic Resolution Crystal Structures Of Kallikrein-Related Peptidase 4 Complexed With Sunflower Trypsin Inhibitor (Sfti-1)
Organism: Homo sapiens, Helianthus annuus
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2014-04-23 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
Atomic Resolution Crystal Structures Of Kallikrein-Related Peptidase 4 Complexed With A Modified Sfti Inhibitor Fcqr
Organism: Homo sapiens, Helianthus annuus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2014-04-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: MPD, LI |
 |
Crystal Structure Of Lipl32, The Most Abundant Surface Protein Of Pathogenic Leptospira Spp
Organism: Leptospira interrogans
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2009-02-24 Classification: UNKNOWN FUNCTION |

