Search Count: 97
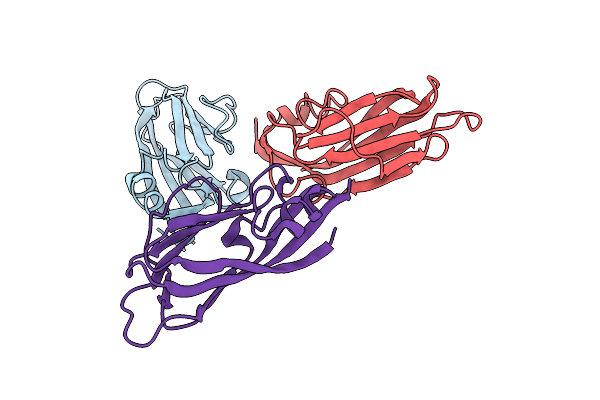 |
Cryoem Structure Of Modified Turnip Yellows Virus Devoid Of Minor Capsid Protein Readthrough Domain
Organism: Turnip yellows virus
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: VIRUS |
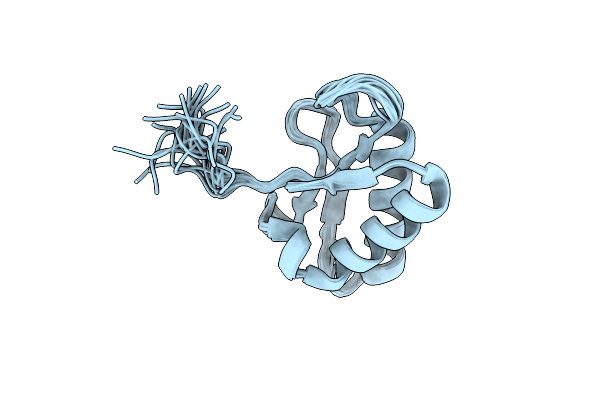 |
Organism: Zymoseptoria tritici st99ch_1e4
Method: SOLUTION NMR Release Date: 2024-10-09 Classification: UNKNOWN FUNCTION |
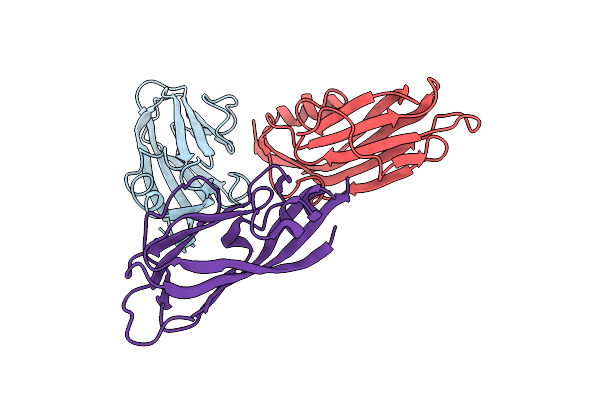 |
Organism: Turnip yellows virus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: VIRUS |
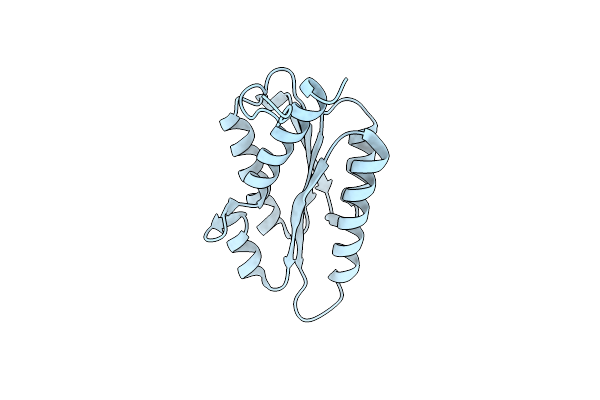 |
Organism: Vicia faba
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: TRANSFERASE |
 |
Organism: Pyricularia oryzae 70-15
Method: SOLUTION NMR Release Date: 2024-02-14 Classification: UNKNOWN FUNCTION |
 |
Organism: Zymoseptoria tritici
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-01-17 Classification: UNKNOWN FUNCTION |
 |
The Crystal Structure Of A Cobalt-Bound Scfv Reveals A Tetrameric Polyhistidine Motif (Tetrhis)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-08-09 Classification: METAL BINDING PROTEIN Ligands: CO, SO4, CXS |
 |
Organism: Thermococcus barophilus
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2023-07-12 Classification: GENE REGULATION |
 |
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2023-07-05 Classification: GENE REGULATION |
 |
Organism: Pyricularia oryzae
Method: SOLUTION NMR Release Date: 2023-04-26 Classification: UNKNOWN FUNCTION |
 |
Organism: Pyricularia oryzae
Method: SOLUTION NMR Release Date: 2023-04-26 Classification: UNKNOWN FUNCTION |
 |
Organism: Pyricularia oryzae
Method: SOLUTION NMR Release Date: 2023-04-26 Classification: UNKNOWN FUNCTION |
 |
Thermococcus Barophilus Phosphomannose Isomerase Protein Structure At 1.6 A
Organism: Thermococcus barophilus
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2022-08-24 Classification: ISOMERASE Ligands: MG |
 |
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2022-08-24 Classification: ISOMERASE Ligands: ZN |
 |
Crystal Structure Of Leukotoxin Luke From Staphylococcus Aureus At 1.9 Angstrom Resolution
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-04-06 Classification: TOXIN Ligands: P6G, MHA, SO4 |
 |
Crystal Structure Of Leukotoxin Luke From Staphylococcus Aureus At 1.5 Angstrom Resolution
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2022-04-06 Classification: TOXIN Ligands: CL |
 |
Crystal Structure Of Leukotoxin Luke From Staphylococcus Aureus In Complex With P-Cresyl Sulfate
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-04-06 Classification: TOXIN Ligands: PEG, IMD, SO4, 6EI |
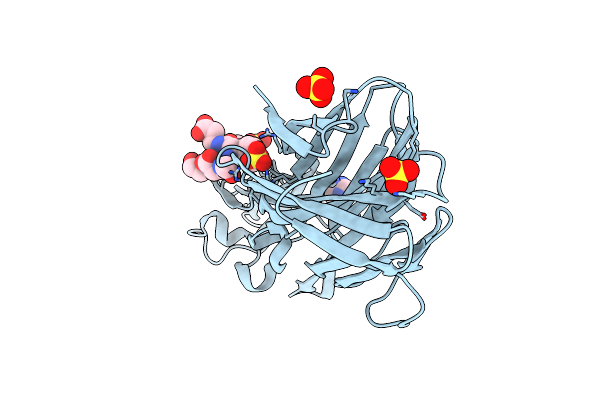 |
Crystal Structure Of Leukotoxin Luke From Staphylococcus Aureus In Complex With A Doubly Sulfated Ccr2 N-Terminal Peptide
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-04-06 Classification: TOXIN Ligands: IMD, SO4 |
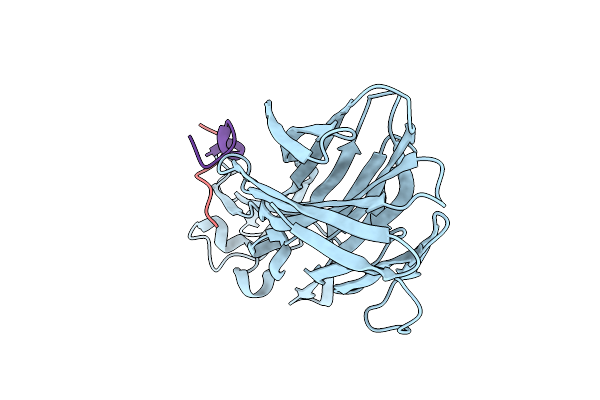 |
Crystal Structure Of Leukotoxin Luke From Staphylococcus Aureus In Complex With A Sulfated Ackr1 N-Terminal Peptide
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-04-06 Classification: TOXIN |
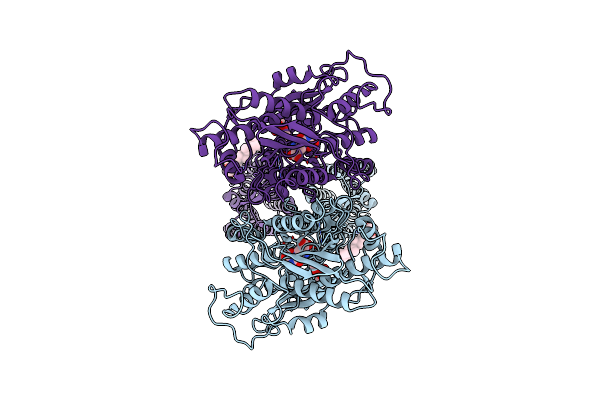 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-08 Classification: MEMBRANE PROTEIN Ligands: NAG, QUS, Y01 |

