Search Count: 4
 |
Structure Of Pcryo_0615 From Psychrobacter Cryohalolentis, An N-Acetyltransferase Required To Produce Diacetamido-2,3-Dideoxy-D-Glucuronic Acid
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-11-23 Classification: TRANSFERASE Ligands: MJZ, COA, NA |
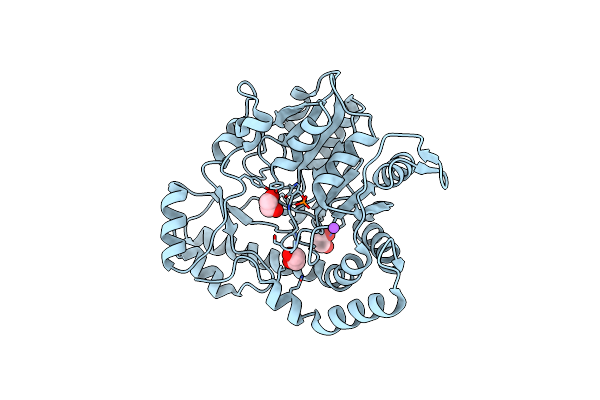 |
Crystal Structure Of Pcryo_0616, The Aminotransferase Required To Synthesize Udp-N-Acetyl-3-Amino-D-Glucosaminuronic Acid (Udp-Glcnac3Na)
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2022-11-23 Classification: TRANSFERASE |
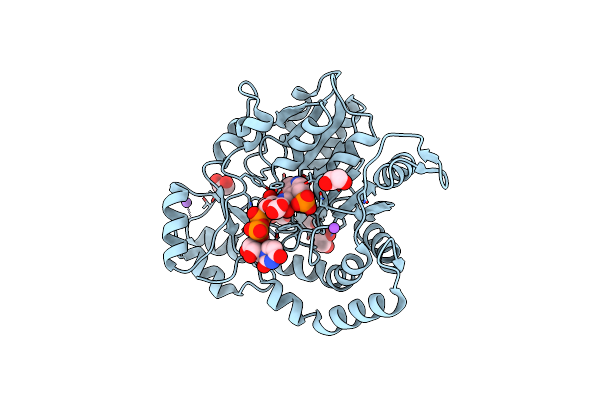 |
Rystal Structure Of Pcryo_0616, The Aminotransferase Required To Synthesize Udp-N-Acetyl-3-Amino-D-Glucosaminuronic Acid (Udp-Glcnac3Na), Incomplete With Its External Aldimine Reaction Intermediate
Organism: Psychrobacter cryohalolentis k5
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2022-11-23 Classification: TRANSFERASE Ligands: ULP, EDO, NA |
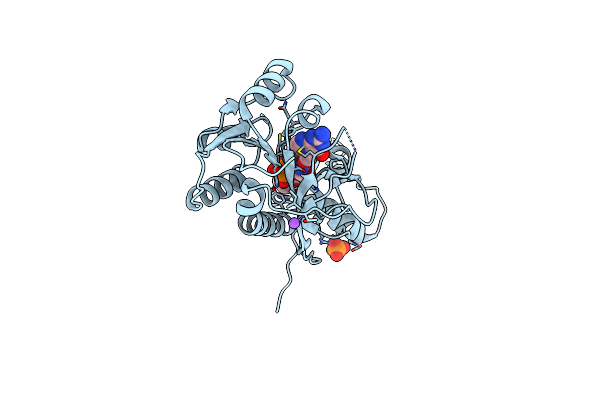 |
Crystal Structure Of A Sugar N-Formyltransferase From The Plant Pathogen Pantoea Ananatis
Organism: Pantoea ananatis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-12-19 Classification: TRANSFERASE Ligands: 0FX, FON, NA, PO4 |

