Search Count: 53
 |
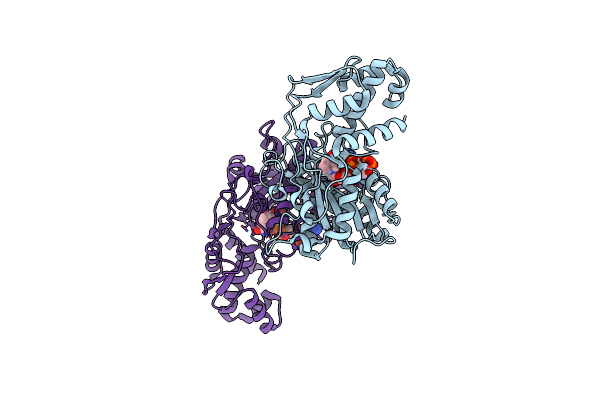
Crystal Structure Of The Intact Mtase From Vibrio Vulnificus Yj016 In Complex With The Dna-Mimicking Ocr Protein And The S-Adenosyl-L-Homocysteine (Sah)
Organism: Vibrio vulnificus (strain yj016), Escherichia phage t7
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-03-23 Classification: TRANSFERASE Ligands: SAH |
 |
Crystal Structure Of A Gdp-6-Ome-4-Keto-L-Xylo-Heptose Reductase From C.Jejuni
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2021-04-14 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
Crystal Structure Of Udp-N-Acetylmuramic Acid L-Alanine Ligase (Murc) From Mycobacterium Bovis
Organism: Mycobacterium bovis (strain atcc baa-935 / af2122/97)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-04-22 Classification: LIGASE Ligands: ZN |
 |
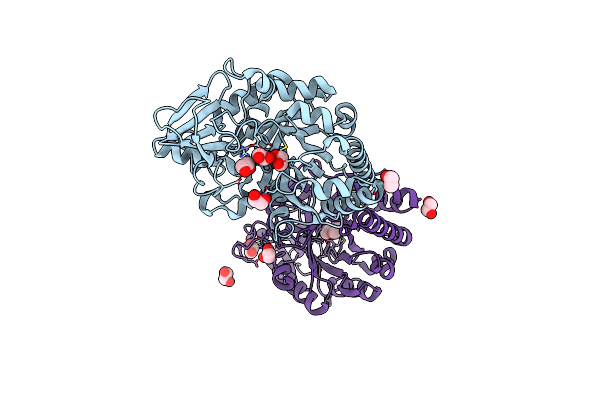
Crystal Structure Of Udp-N-Acetylmuramic Acid L-Alanine Ligase (Murc) From Mycobacterium Bovis In Complex With Udp-N-Acetylglucosamine
Organism: Mycobacterium bovis (strain atcc baa-935 / af2122/97)
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2020-04-22 Classification: LIGASE Ligands: UD1, ZN |
 |
Crystal Structure Of Immunomodulatory Active Chitinase From Trichuris Suis, Tses1
Organism: Trichuris suis
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2019-04-24 Classification: HYDROLASE Ligands: EDO |
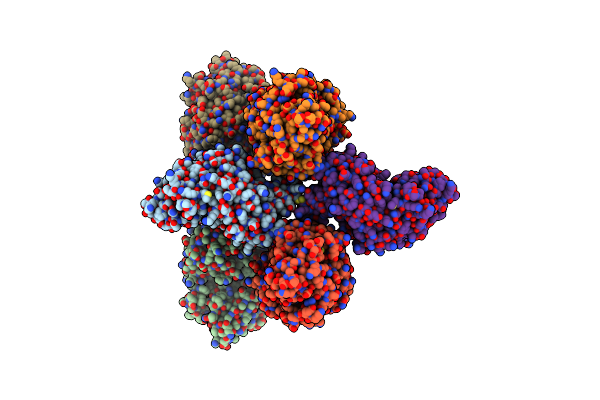 |
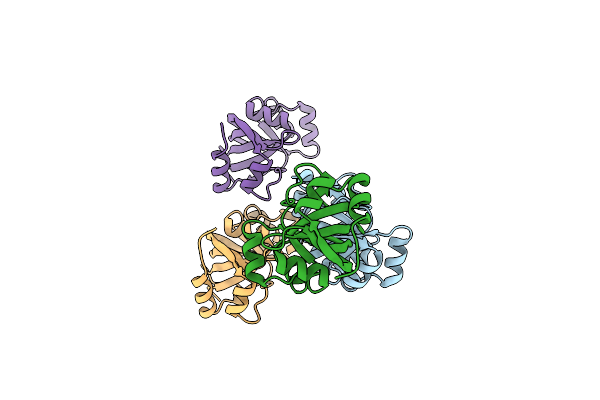
Crystal Structure Of Immunomodulatory Active Chitinase From Trichuris Suis - Tses1 - 6 Molecules In Asu
Organism: Trichuris suis
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2019-04-24 Classification: HYDROLASE Ligands: EDO |
 |
Organism: Zymomonas mobilis subsp. mobilis (strain atcc 10988 / dsm 424 / lmg 404 / ncimb 8938 / nrrl b-806 / zm1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-07-18 Classification: RNA BINDING PROTEIN |
 |
Organism: Zymomonas mobilis subsp. mobilis (strain atcc 10988 / dsm 424 / lmg 404 / ncimb 8938 / nrrl b-806 / zm1)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-07-18 Classification: RNA BINDING PROTEIN Ligands: CL |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-05-09 Classification: LYASE Ligands: MG, CO3 |
 |
Organism: Pseudomonas sp. hmsc75e02
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-05-09 Classification: LYASE Ligands: ACY, EDO, NI |
 |
Organism: Pseudomonas sp. hmsc75e02
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-05-09 Classification: LYASE Ligands: CA, ACY, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2017-12-13 Classification: HYDROLASE Ligands: ZN, ACT, YB, IMD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-12-13 Classification: HYDROLASE Ligands: ZN, ACT, YB, IMD, BGW |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2017-12-13 Classification: HYDROLASE Ligands: ZN, ACT, YB, IMD, BGK |
 |
Organism: Zymomonas mobilis subsp. mobilis (strain atcc 10988 / dsm 424 / lmg 404 / ncimb 8938 / nrrl b-806 / zm1)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-08-30 Classification: HYDROLASE |
 |
Organism: Zymomonas mobilis subsp. mobilis (strain atcc 10988 / dsm 424 / lmg 404 / ncimb 8938 / nrrl b-806 / zm1)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-08-30 Classification: HYDROLASE Ligands: PE8, CL, SO4 |
 |
Crystal Structure Of Rv1284 In The Presence Of Polycarpine At Mildly Acidic Ph
Organism: Mycobacterium tuberculosis (strain cdc 1551 / oshkosh)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-05-06 Classification: LYASE Ligands: ZN, MG, CL |
 |
Organism: Mycobacterium tuberculosis (strain cdc 1551 / oshkosh)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-05-06 Classification: LYASE Ligands: ZN, CL |
 |
Organism: Mycobacterium tuberculosis (strain cdc 1551 / oshkosh)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2015-05-06 Classification: LYASE Ligands: ZN, MG, CL |
 |
Organism: Necator americanus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-05-14 Classification: UNKNOWN FUNCTION Ligands: CO |

