Search Count: 56
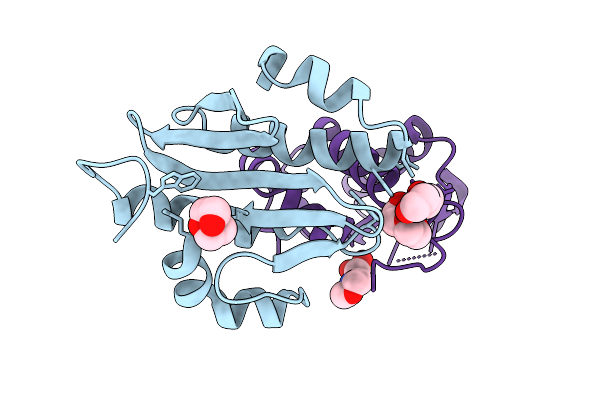 |
Organism: Listeria monocytogenes 10403s
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: METAL BINDING PROTEIN Ligands: PEG, MES |
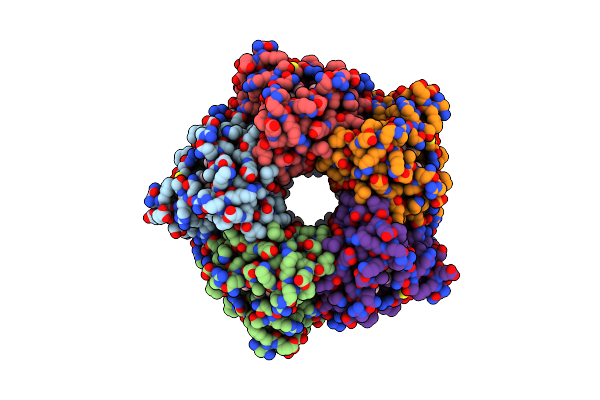 |
Cryo-Em Structure Of Coproheme Decarboxylase From Corynebacterium Diphtheriae In Complex With Heme B
Organism: Corynebacterium diphtheriae
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: HEM |
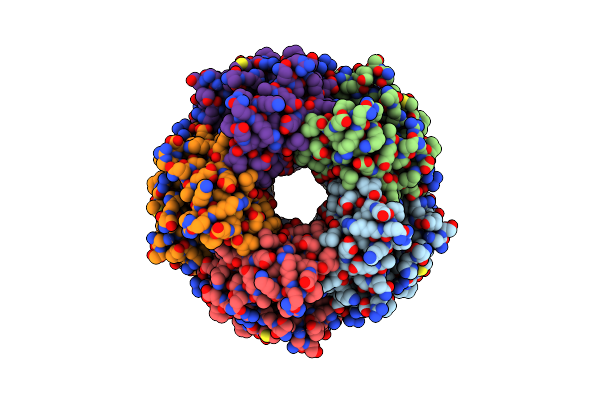 |
Cryo-Em Structure Of Apo Coproheme Decarboxylase From Corynebacterium Diphtheria.
Organism: Corynebacterium diphtheriae
Method: ELECTRON MICROSCOPY Resolution:2.27 Å Release Date: 2025-02-05 Classification: OXIDOREDUCTASE |
 |
Coproporphyrin Iii - Lmcpfc Wt Complex Soaked With Fe2+ And Anomalous Densities
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-04-24 Classification: METAL BINDING PROTEIN Ligands: HT9, EDO, FE, CL |
 |
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-04-24 Classification: METAL BINDING PROTEIN Ligands: FEC, EDO, CA |
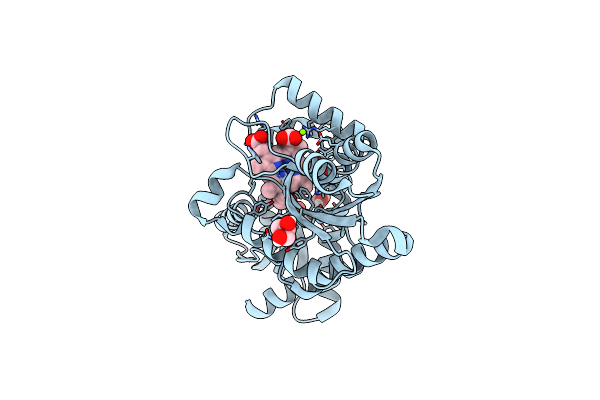 |
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-05-03 Classification: METAL BINDING PROTEIN Ligands: GOL, EDO, HT9, FE2, MG |
 |
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-04-05 Classification: METAL BINDING PROTEIN Ligands: GOL, ACT, HT9, EDO, FE2 |
 |
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2023-01-11 Classification: METAL BINDING PROTEIN Ligands: GOL, EDO, HT9, FE2, CL |
 |
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2022-12-28 Classification: METAL BINDING PROTEIN Ligands: ACT, GOL, EDO, HT9, CA |
 |
Organism: Listeria monocytogenes
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2022-12-28 Classification: METAL BINDING PROTEIN Ligands: GOL, HT9 |
 |
Structure Of Native Leukocyte Myeloperoxidase In Complex With The Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2022-10-26 Classification: OXIDOREDUCTASE Ligands: EDO, HEC, CL, CA, PO4, PEG |
 |
Structure Of Native Leukocyte Myeloperoxidase In Complex With A Truncated Version (Spin Truncated) Of The Staphyloccal Peroxidase Inhibitor Spin From Staphylococcus Aureus
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2022-10-26 Classification: OXIDOREDUCTASE Ligands: CL, HEC, CA, EDO, NAG, ACT, OXL |
 |
Structure Of Coproheme Decarboxylase From Corynebacterium Dipththeriae W183Y Mutant In Complex With Coproheme
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-02-23 Classification: BIOSYNTHETIC PROTEIN Ligands: FEC, PGE, PEG |
 |
Structure Of Coproheme Decarboxylase From Corynebacterium Dipththeriae Y135A Mutant In Complex With Coproheme
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2022-02-23 Classification: BIOSYNTHETIC PROTEIN Ligands: FEC, PEG |
 |
Crystal Structure Of Dimeric Chlorite Dismutase From Cyanothece Sp. Pcc7425 In Complex With Nitrite
Organism: Cyanothece sp. (strain pcc 7425 / atcc 29141)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-12-22 Classification: OXIDOREDUCTASE Ligands: NO2, HEM, GOL, SO4 |
 |
Crystal Structure Of Dimeric Chlorite Dismutase Variant Q74V (Ccld Q74V) From Cyanothece Sp. Pcc7425 In Complex With Nitrite
Organism: Cyanothece sp. (strain pcc 7425 / atcc 29141)
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2021-12-22 Classification: OXIDOREDUCTASE Ligands: HEM, NO2, GOL |
 |
Crystal Structure Of Dimeric Chlorite Dismutase Variant Q74E (Ccld Q74E) From Cyanothece Sp. Pcc7425 In Complex With Nitrite
Organism: Cyanothece sp. (strain pcc 7425 / atcc 29141)
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2021-12-22 Classification: OXIDOREDUCTASE Ligands: HEM, NO2, SO4, GOL, PGE |
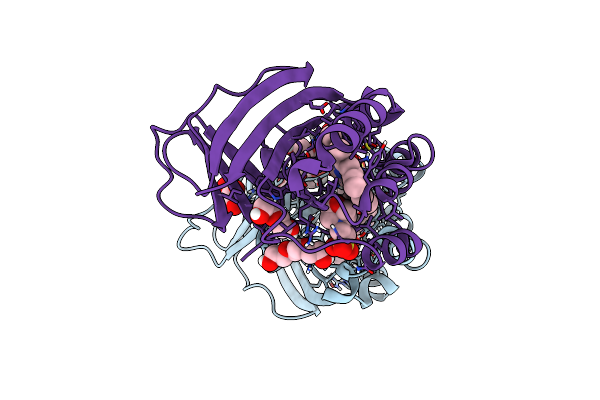 |
Crystal Structure Of Dimeric Chlorite Dismutase Variant R127K (Ccld R127K) From Cyanothece Sp. Pcc7425
Organism: Cyanothece sp. (strain pcc 7425 / atcc 29141)
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2021-12-22 Classification: OXIDOREDUCTASE Ligands: HEM, GOL, MES, PGE, SO4, PEG |
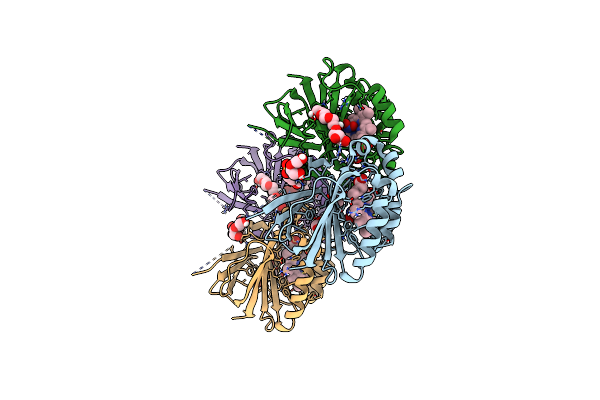 |
Crystal Structure Of Dimeric Chlorite Dismutase Variant R127A (Ccld R127A) From Cyanothece Sp. Pcc7425 In Complex With Nitrite
Organism: Cyanothece sp. pcc 7425
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-12-22 Classification: OXIDOREDUCTASE Ligands: PGE, GOL, HEM, NO2, PEG, SO4 |
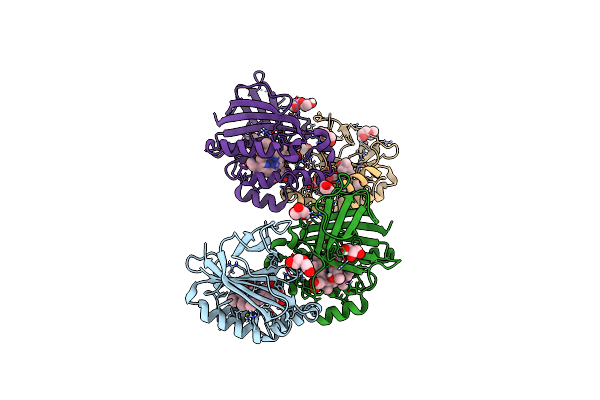 |
Crystal Structure Of Dimeric Chlorite Dismutase Variant R127A (Ccld R127A) From Cyanothece Sp. Pcc7425
Organism: Cyanothece sp. pcc 7425
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-12-22 Classification: OXIDOREDUCTASE Ligands: HEM, PEG, MPD, GOL |

