Search Count: 49
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: DNA BINDING PROTEIN Ligands: MG |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: SHT, TP7, COM, F43, S5Q, ZN, ATP, MG |
 |
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: F43, SHT, COM, TP7, S5Q |
 |
Methyl-Coenzyme M Reductase Activation Complex Binding To The A2 Component After Incubation With Atp
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: F43, COM, TP7, SHT, S5Q, ZN, ATP, MG |
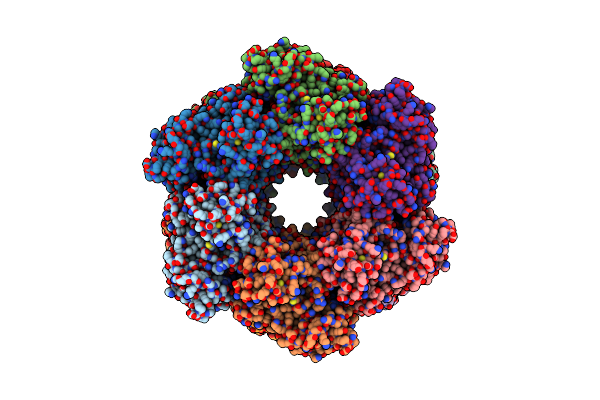 |
Cryo-Em Structure Of The Active Dodecameric Methanosarcina Mazei Glutamine Synthetase.
Organism: Methanosarcina mazei go1
Method: ELECTRON MICROSCOPY Release Date: 2025-01-08 Classification: CYTOSOLIC PROTEIN Ligands: AKG |
 |
Periplasmic Scaffold Of The Campylobacter Jejuni Flagellar Motor (Alpha Carbon Trace)
Organism: Campylobacter jejuni
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: STRUCTURAL PROTEIN |
 |
Organism: Klebsiella pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: GENE REGULATION Ligands: ZN |
 |
Organism: Pseudomonas oleovorans
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: GENE REGULATION Ligands: ZN |
 |
Dna Bound Type Iv-A1 Crispr Effector Complex With The Ding Helicase From P. Oleovorans
Organism: Pseudomonas oleovorans
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: GENE REGULATION Ligands: ZN |
 |
Dna-Bound Type Iv-A3 Crispr Effector In Complex With Ding Helicase From K. Pneumoniae (State I)
Organism: Klebsiella pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ANTIVIRAL PROTEIN Ligands: ZN |
 |
Dna-Bound Type Iv-A3 Crispr Effector In Complex With Ding Helicase From K. Pneumoniae (State Ii)
Organism: Klebsiella pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ANTIVIRAL PROTEIN Ligands: ZN |
 |
Dna-Bound Type Iv-A3 Crispr Effector In Complex With Ding Helicase From K. Pneumoniae (State Iii)
Organism: Klebsiella pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ANTIVIRAL PROTEIN Ligands: ZN |
 |
Cvhsp (Hspb7) C131S Alpha-Crystallin Domain - Filamin C (Flnc) Domain 24 Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2024-10-16 Classification: CHAPERONE |
 |
L2 Forming Rubisco Derived From Ancestral Sequence Reconstruction Of The Last Common Ancestor Of Form I'' And Form I Rubiscos
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-10-16 Classification: LYASE Ligands: CAP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2024-10-16 Classification: CHAPERONE |
 |
Non-Obligately L8S8-Complex Forming Rubisco Derived From Ancestral Sequence Reconstruction And Rational Engineering In L8S8 Complex With Substitutions R269W, E271R, L273N
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-10-02 Classification: LYASE Ligands: CAP, MG, B3P |
 |
Organism: Methylophaga sulfidovorans
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-07-31 Classification: TRANSFERASE |
 |
Organism: Ananas comosus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: TRANSFERASE |

