Search Count: 22
 |
Organism: Rhodobacteraceae bacterium qy30, Dna molecule
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: ANTIVIRAL PROTEIN/DNA Ligands: MN |
 |
Organism: Rhodobacteraceae bacterium qy30
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: ANTIVIRAL PROTEIN/DNA Ligands: PO4 |
 |
Organism: Rhodobacteraceae bacterium qy30
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: ANTIVIRAL PROTEIN Ligands: A1A3G, F2A, MN, A1A3H, D5M |
 |
Organism: Rhodobacteraceae bacterium qy30, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: ANTIVIRAL PROTEIN |
 |
Structure Of Bacillus Phage Spo1 Anti-Cbass 4 (Acb4) In Complex With 3'3'-Cgamp
Organism: Bacillus phage spo1
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2025-01-29 Classification: VIRAL PROTEIN Ligands: 4BW |
 |
Organism: Bacillus cereus, Metagenome
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-22 Classification: VIRAL PROTEIN |
 |
Organism: Penguinpox virus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-09-04 Classification: VIRAL PROTEIN Ligands: A1AQN, GUN |
 |
Organism: Penguinpox virus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-09-04 Classification: VIRAL PROTEIN Ligands: A1AWU |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2024-06-19 Classification: UNKNOWN FUNCTION |
 |
Structure Of Clostridium Botulinum Prophage Tad1 In Complex With 1''-3' Gcadpr
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-11-22 Classification: UNKNOWN FUNCTION Ligands: OJC |
 |
Organism: Okubovirus
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2023-11-22 Classification: VIRAL PROTEIN |
 |
Organism: Okubovirus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-11-22 Classification: VIRAL PROTEIN Ligands: OJC, MG |
 |
Organism: Okubovirus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-11-22 Classification: VIRAL PROTEIN Ligands: MF6 |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-11-22 Classification: VIRAL PROTEIN |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-09-20 Classification: MEMBRANE PROTEIN |
 |
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-10-05 Classification: VIRAL PROTEIN |
 |
Structure Of Clostridium Botulinum Prophage Tad1 In Complex With 1''-2' Gcadpr
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2022-10-05 Classification: VIRAL PROTEIN Ligands: MF6 |
 |
Organism: Erwinia phage fbb1
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2022-04-20 Classification: VIRAL PROTEIN |
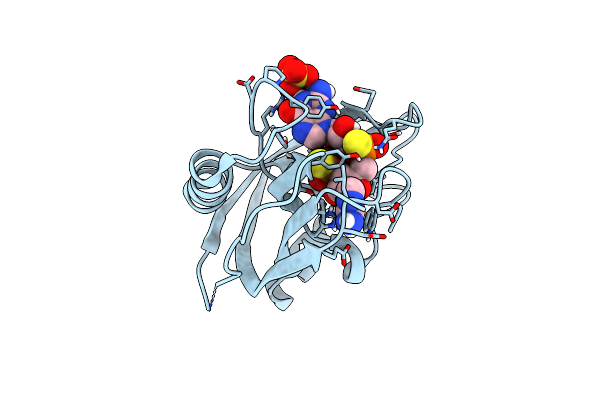 |
Structure Of Phage Fbb1 Anti-Cbass Nuclease Acb1-3'3'-Cgamp Complex In Post Reaction State
Organism: Erwinia phage fbb1
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-04-20 Classification: VIRAL PROTEIN Ligands: ECI, SO4 |
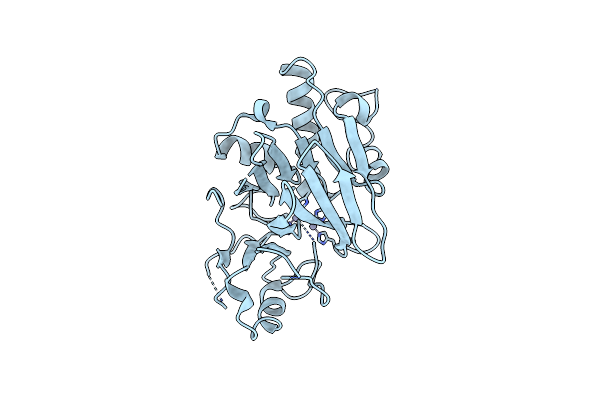 |
Organism: Bacillus phage bsp38
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2022-04-20 Classification: VIRAL PROTEIN Ligands: ZN |

