Search Count: 169
 |
Crystal Structure Of Acyl-Coa Dehydrogenase Fade2 Mutant (Pa0508 M130G E296A Q303A) From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2025-02-19 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, PGE |
 |
Crystal Structure Of The Acyl-Coa Dehydrogenase Fade1(Pa0506) E441A From Pseudomonas Aeruginosa Complexed With C16Coa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: PKZ, EDO, PGE, NO3, TRS |
 |
Crystal Structure Of The Apo Acyl-Coa Dehydrogenase Fade2 (Pa0508) From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, 1PE, GOL |
 |
Crystal Structure Of The Acyl-Coa Dehydrogenase Pa0506 (Fade1) From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, CO8 |
 |
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-11-06 Classification: RNA BINDING PROTEIN |
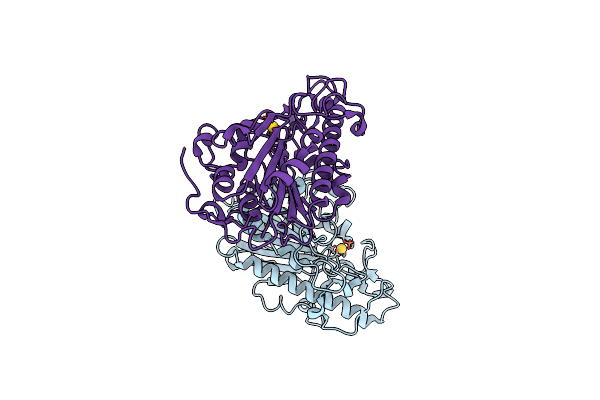 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2023-02-01 Classification: TRANSFERASE Ligands: AU |
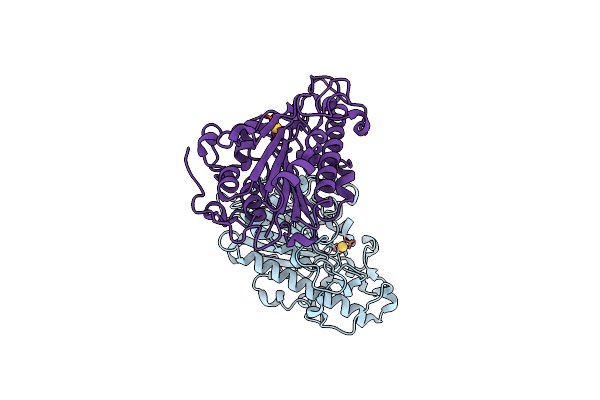 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-02-01 Classification: TRANSFERASE Ligands: AU |
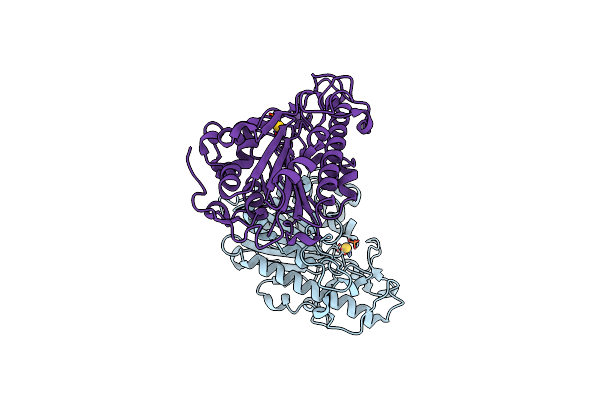 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-02-01 Classification: TRANSFERASE Ligands: AU |
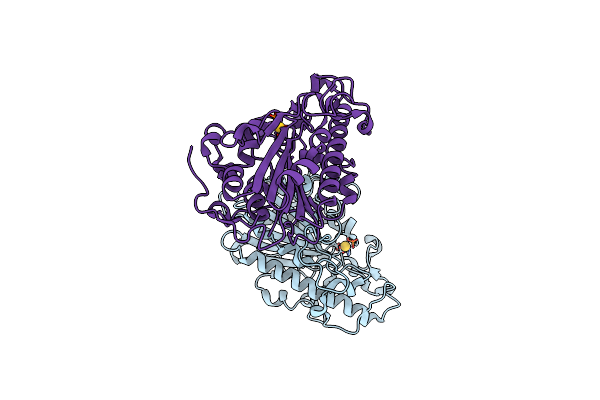 |
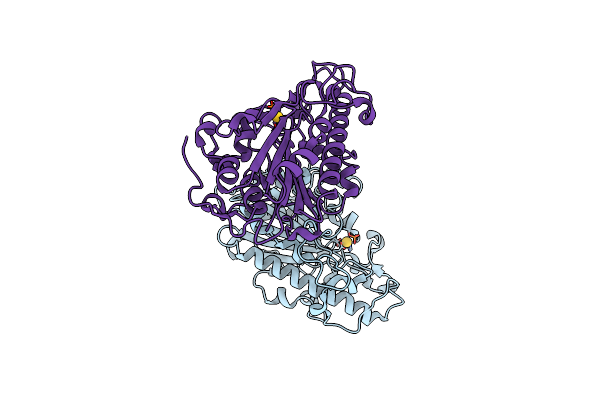
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-02-01 Classification: TRANSFERASE Ligands: AU |
 |
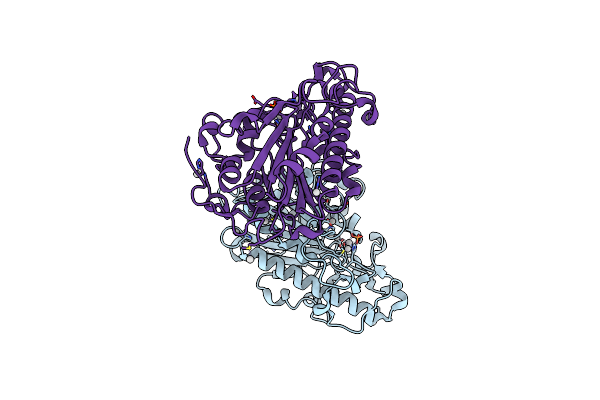
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-02-01 Classification: TRANSFERASE Ligands: AU |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2022-03-16 Classification: TRANSFERASE Ligands: AG |
 |
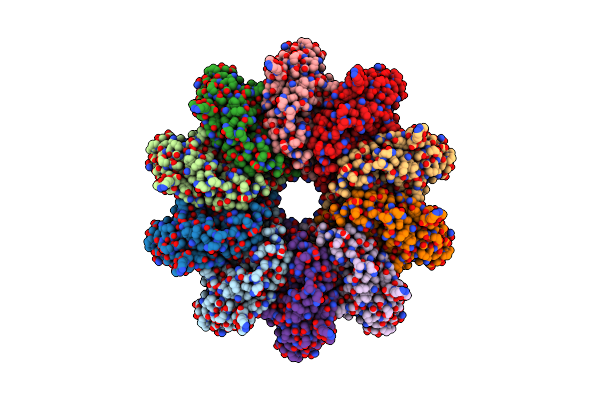
Organism: Homo sapiens, Human gammaherpesvirus 4
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-01-12 Classification: IMMUNE SYSTEM Ligands: CL, GOL |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2021-10-13 Classification: MEMBRANE PROTEIN |
 |
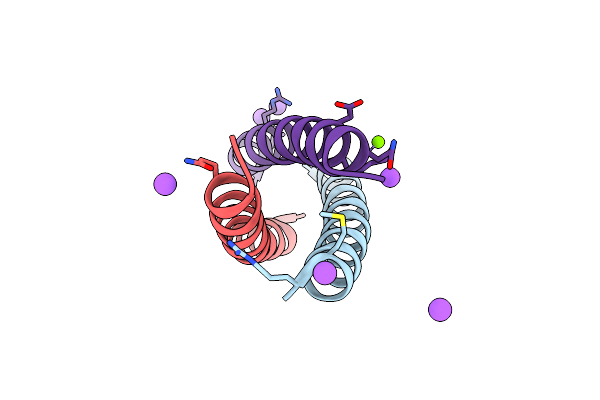
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2021-10-13 Classification: MEMBRANE PROTEIN |
 |
Gcn4-P1 Peptide Trimer With P-Methylphenylalanine Residue At Position 16 (Me-F16)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2021-07-07 Classification: TRANSCRIPTION |
 |
Gcn4-P1 Peptide Trimer With Tetrafluoroiodophenylalanine Residue At Position 16 (Tfi-F16)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-07-07 Classification: TRANSCRIPTION |
 |
Gcn4-P1 Peptide Trimer With Iodo-Phenylalanine Residue At Position 16 (Ipf-F16)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-07-07 Classification: TRANSCRIPTION |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-07-07 Classification: TRANSCRIPTION |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2020-09-16 Classification: HYDROLASE Ligands: AU, SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-09-16 Classification: TRANSFERASE Ligands: ZN |

