Search Count: 54
 |
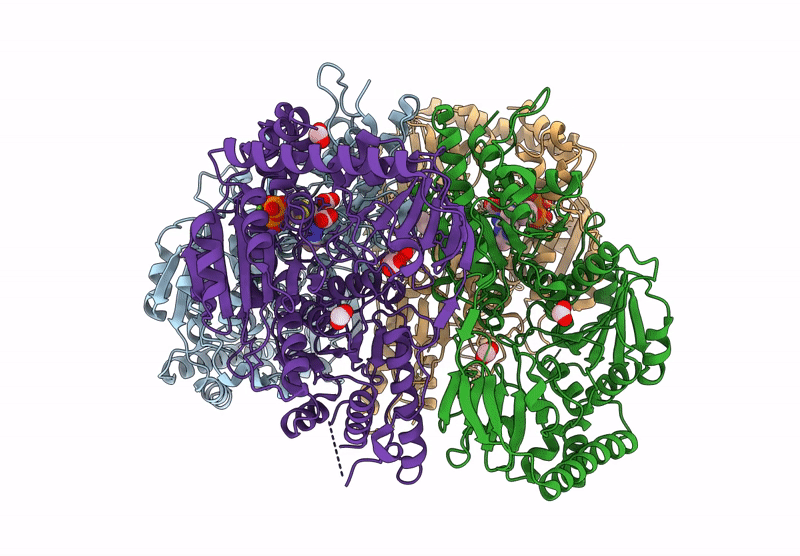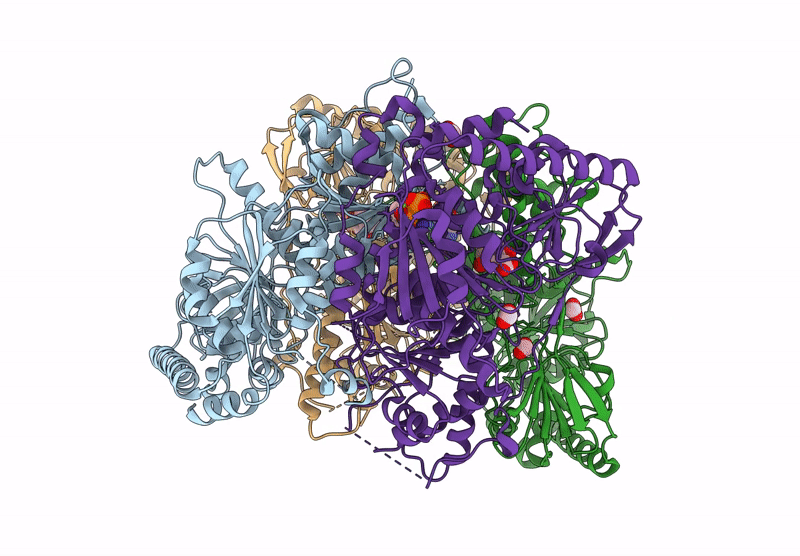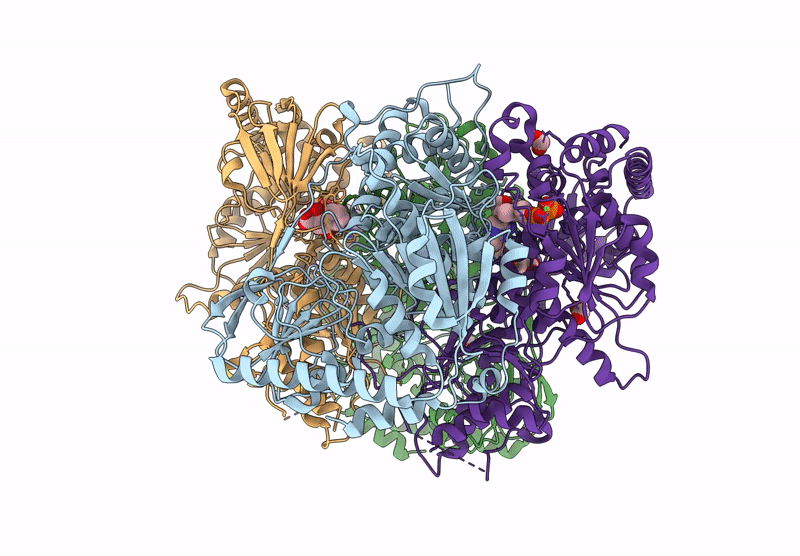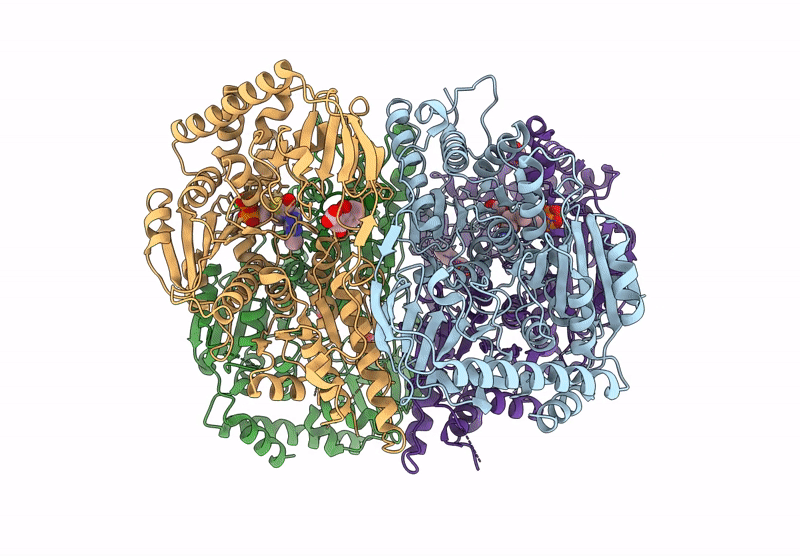
Organism: Mycobacterium tuberculosis h37rv
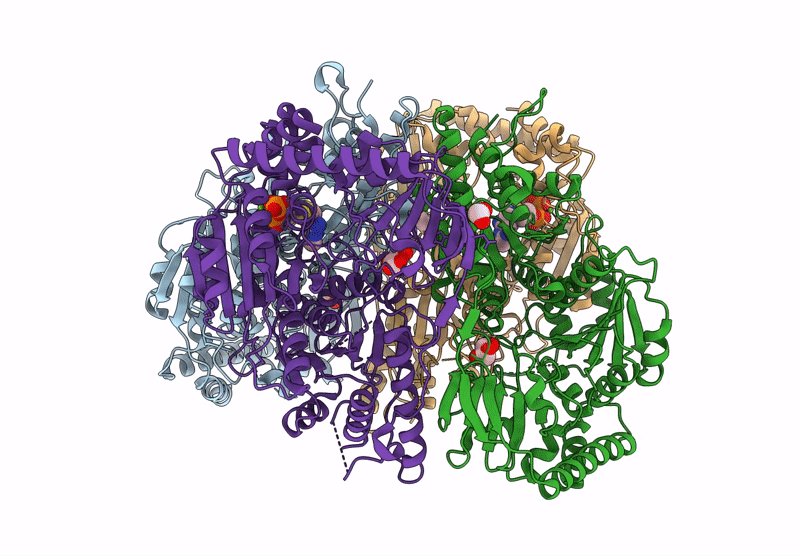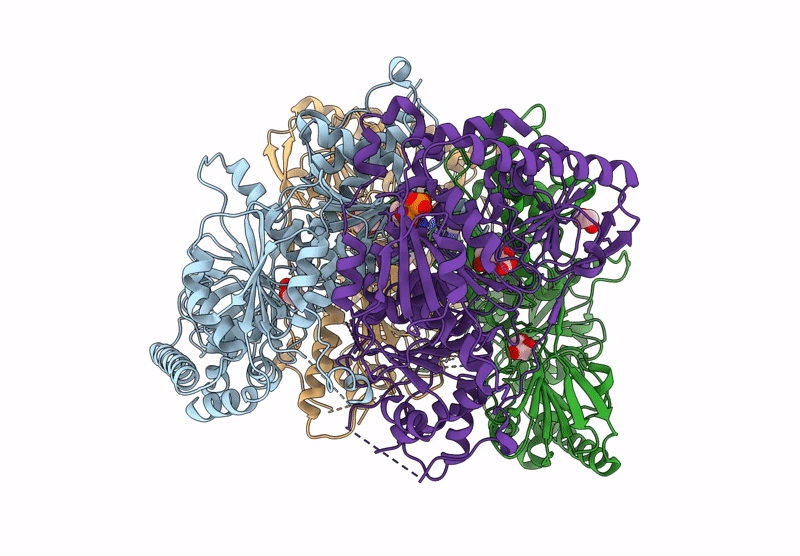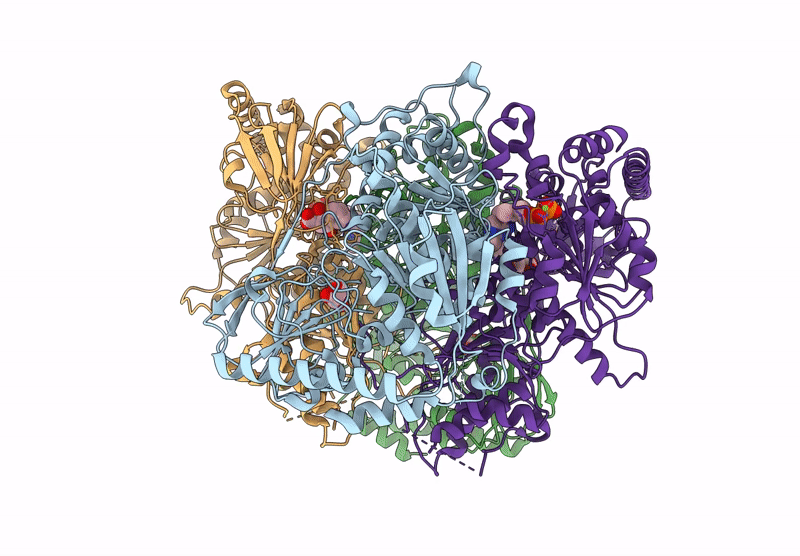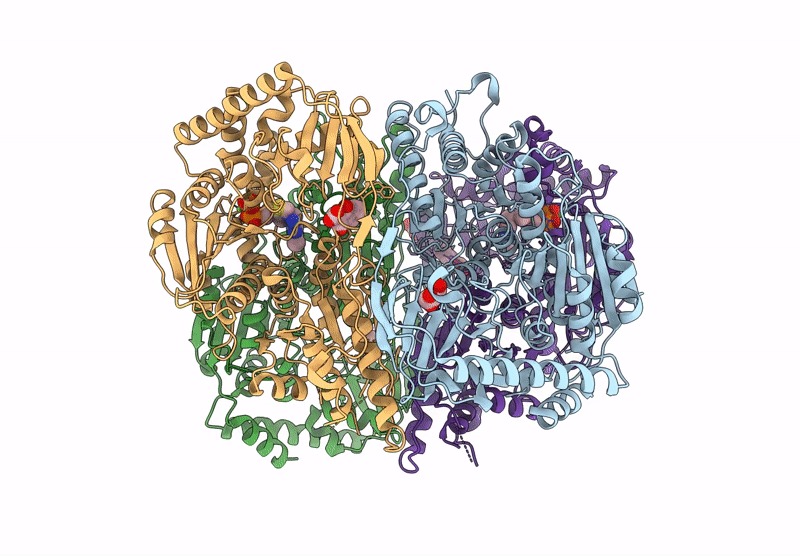
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2025-04-16 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, CL, TPP, DNA, MG |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-04-16 Classification: BIOSYNTHETIC PROTEIN Ligands: FMT, TPP, DNA, MG, CL |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2025-04-16 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, CL, TPP, DNA, EDO, MG |
 |
Crystal Structure Of Plasmodium Falciparum Dihydroorotate Dehydrogenase Bound With Inhibitor Dsm1010 (6-Cyclopropyl-2,4-Dimethyl-3-(4-(Trifluoromethyl)Benzyl)-2,6-Dihydro-7H-Pyrazolo[3,4-C]Pyridin-7-One)
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: A1A5P, FMN, OG6 |
 |
Crystal Structure Of Plasmodium Falciparum Dihydroorotate Dehydrogenase Bound With Inhibitor Dsm679 (Ethyl 1,4-Dimethyl-5-((6-(Trifluoromethyl)Pyridin-3-Yl)Methyl)-1H-Pyrazole-3-Carboxylate)
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2025-01-01 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: A1A51, FMN, OG6, CIT, LDA |
 |
Crystal Structure Of Plasmodium Falciparum Dihydroorotate Dehydrogenase Bound With Inhibitor Dsm681 (N-Cyclopropyl-1,4-Dimethyl-5-((6-(Trifluoromethyl)Pyridin-3-Yl)Methyl)-1H-Pyrazole-3-Carboxamide)
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-01-01 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: A1A52, FMN, OG6, NA |
 |
Crystal Structure Of Plasmodium Falciparum Dihydroorotate Dehydrogenase Bound With Inhibitor Dsm959 (6-Cyclopropyl-2,4-Dimethyl-3-((6-(Trifluoromethyl)Pyridin-3-Yl)Methyl)-2,6-Dihydro-7H-Pyrazolo[3,4-C]Pyridin-7-One)
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-01-01 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: A1A53, FMN, OG6 |
 |
Crystal Structure Of Plasmodium Falciparum Dihydroorotate Dehydrogenase Bound With Inhibitor Dsm1153 (4,6-Dicyclopropyl-3-(3-Fluoro-4-(Trifluoromethyl)Benzyl)-2-Methyl-2,6-Dihydro-7H-Pyrazolo[3,4-D]Pyridazin-7-One)
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2025-01-01 Classification: OXIDOREDUCTASE Ligands: A1A5Q, FMN, OG6 |
 |
Crystal Structure Of Plasmodium Falciparum Dihydroorotate Dehydrogenase Bound With Inhibitor Dsm1174 (3-((7-Azaspiro[3.5]Nonan-7-Yl)Methyl)-4-Cyclopropyl-6-Ethyl-2-Methyl-2,6-Dihydro-7H-Pyrazolo[3,4-C]Pyridin-7-One)
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2025-01-01 Classification: OXIDOREDUCTASE Ligands: A1A59, FMN, OG6 |
 |
Crystal Structure Of Plasmodium Falciparum Dihydroorotate Dehydrogenase Bound With Inhibitor Dsm1398 ((S)-3-(Amino(3-Chloro-4-(Trifluoromethyl)Phenyl)(Cyclopropyl)Methyl)-6-Cyclopropyl-2-Methyl-2,6-Dihydro-7H-Pyrazolo[3,4-D]Pyridazin-7-One)
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2025-01-01 Classification: OXIDOREDUCTASE Ligands: A1A6D, FMN, OG6, ACY |
 |
Crystal Structure Of Plasmodium Falciparum Dihydroorotate Dehydrogenase Bound With Inhibitor Dsm1211 ((R)-3-(Amino(6-(Trifluoromethyl)Pyridin-3-Yl)Methyl)-4-Cyclopropyl-6-Ethyl-2-Methyl-2,6-Dihydro-7H-Pyrazolo[3,4-C]Pyridin-7-One)
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-01-01 Classification: OXIDOREDUCTASE Ligands: A1A6E, FMN, OG6 |
 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: CYTOSOLIC PROTEIN/INHIBITOR Ligands: A1AE6 |
 |
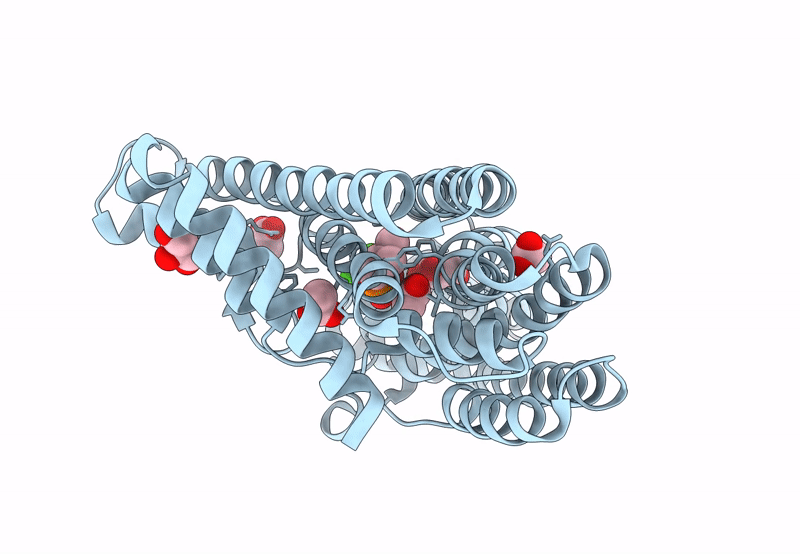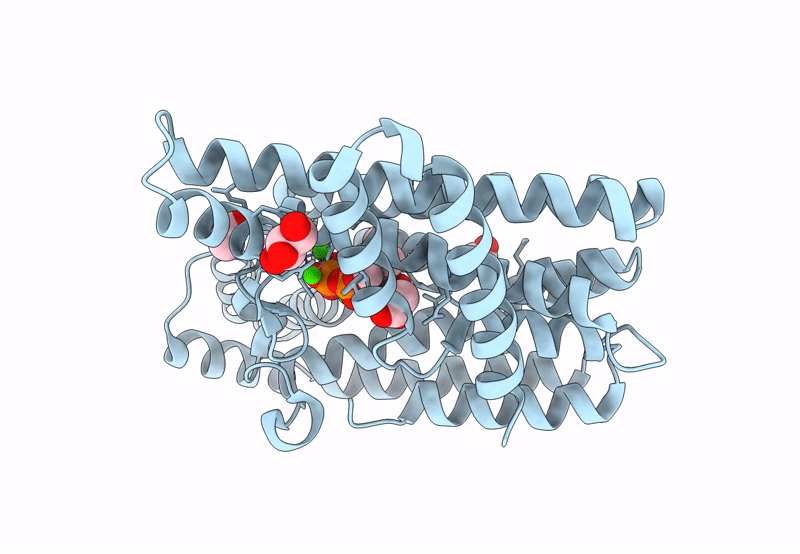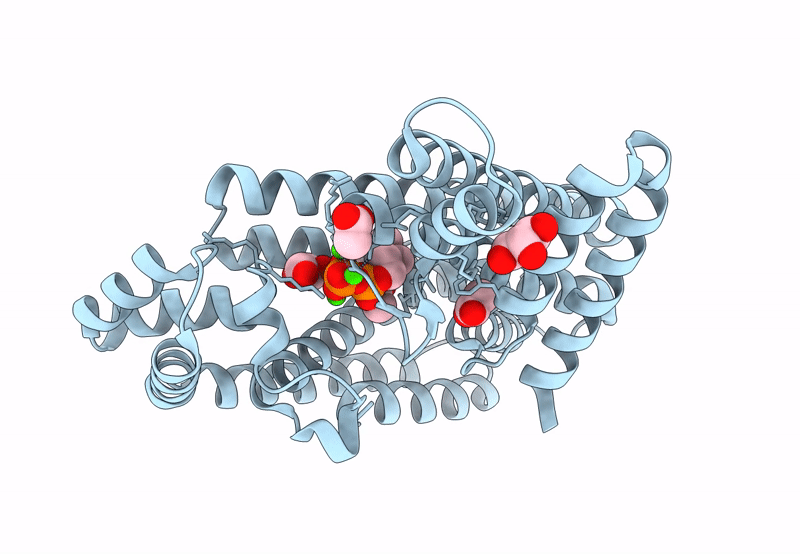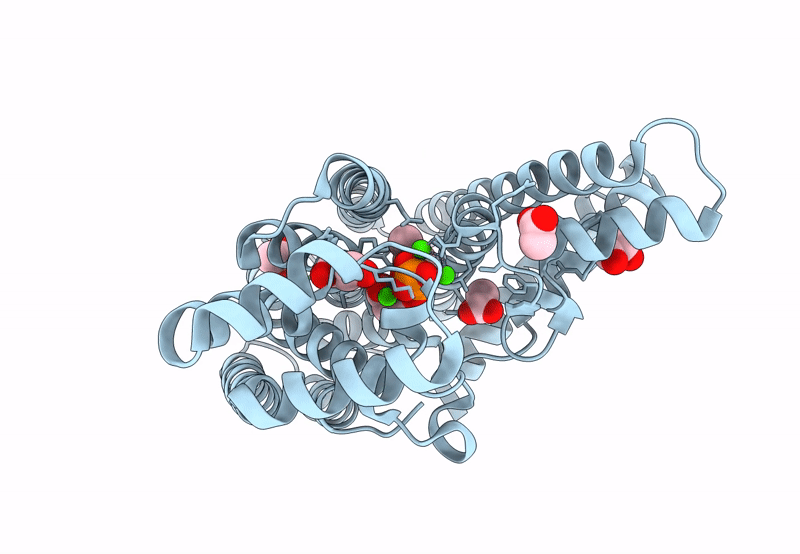
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-11-29 Classification: TRANSFERASE Ligands: GOL, MG |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-11-29 Classification: TRANSFERASE Ligands: GOL, ACT, IPE, CA, CL |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-11-29 Classification: TRANSFERASE Ligands: DMA, GOL, CA |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-09-14 Classification: TRANSFERASE Ligands: TPP, GLY, CA, NA, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-08-17 Classification: METAL BINDING PROTEIN Ligands: 85H, SO4, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2022-08-17 Classification: METAL BINDING PROTEIN Ligands: 85H, CA |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-02-19 Classification: transferase/transferase inhibitor Ligands: DNA, ACT, FMT, MG, TOI, CL |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-02-19 Classification: transferase/transferase inhibitor Ligands: DNA, ACT, FMT, AKG, TOG, MG, CL, PGE, GOL |

