Search Count: 53
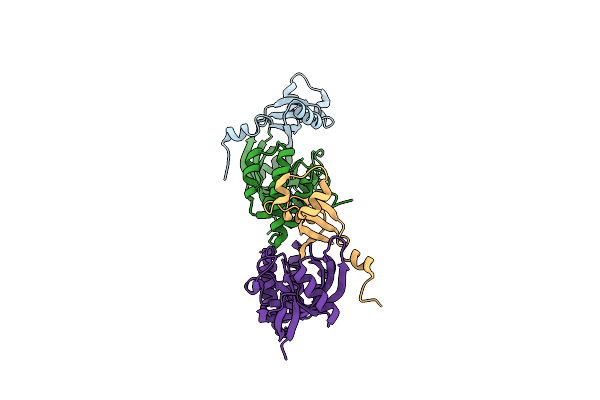 |
Organism: Caulobacter vibrioides (strain atcc 19089 / cip 103742 / cb 15)
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2025-03-12 Classification: HYDROLASE |
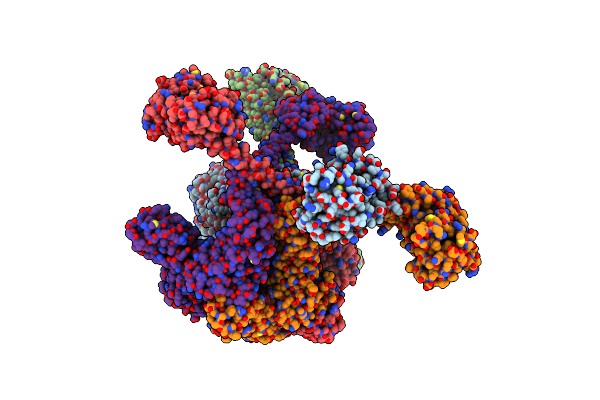 |
Open-Spiral Pentamer Of The Substrate-Free Lon Protease With A Y224S Mutation
Organism: Meiothermus taiwanensis
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: HYDROLASE Ligands: AGS |
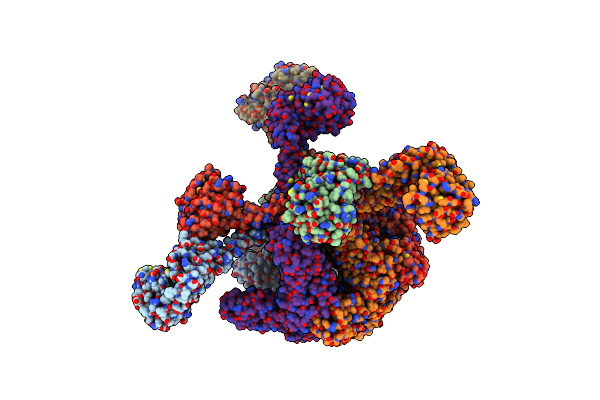 |
Organism: Meiothermus taiwanensis
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: HYDROLASE Ligands: AGS |
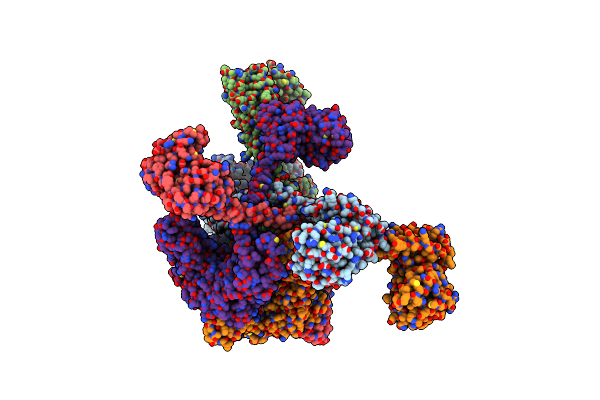 |
Organism: Meiothermus taiwanensis
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: HYDROLASE Ligands: ADP |
 |
Close-Ring Hexamer Of The Substrate-Bound Lon Protease With An S678A Mutation
Organism: Meiothermus taiwanensis, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: HYDROLASE Ligands: ADP |
 |
Organism: Meiothermus taiwanensis
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: HYDROLASE |
 |
Organism: Meiothermus taiwanensis
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: HYDROLASE |
 |
Organism: Meiothermus taiwanensis
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: HYDROLASE |
 |
Organism: Meiothermus taiwanensis
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: HYDROLASE |
 |
Organism: Meiothermus taiwanensis
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: HYDROLASE |
 |
Organism: Meiothermus taiwanensis
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: HYDROLASE Ligands: ADP |
 |
Organism: Meiothermus taiwanensis
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: HYDROLASE Ligands: ADP |
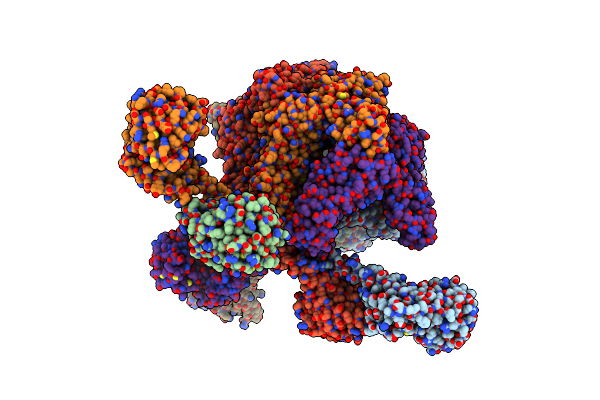 |
Organism: Meiothermus taiwanensis
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: HYDROLASE Ligands: ADP |
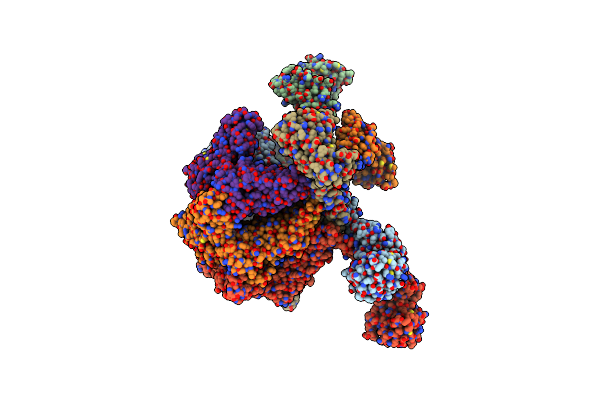 |
The "5+1" Heteromeric Structure Of Lon Protease Consisting Of A Spiral Pentamer With Y224S Mutation And An N-Terminal-Truncated Monomeric E613K Mutant
Organism: Meiothermus taiwanensis
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: HYDROLASE Ligands: ADP |
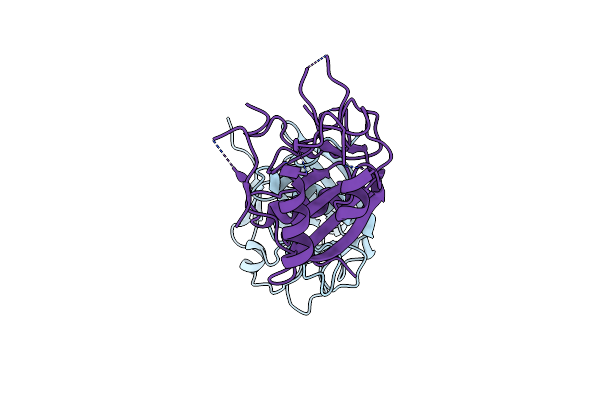 |
Organism: Escherichia coli 908519
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-10-11 Classification: HYDROLASE |
 |
Robust Design Of Effective Allosteric Activator Ubv R4 For Rsp5 E3 Ligase Using The Machine-Learning Tool Proteinmpnn
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-08-09 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4 |
 |
Organism: Escherichia coli dh5[alpha]
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Escherichia coli dh5[alpha]
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-03-29 Classification: BIOSYNTHETIC PROTEIN Ligands: CL, NA, BME, GOL |
 |
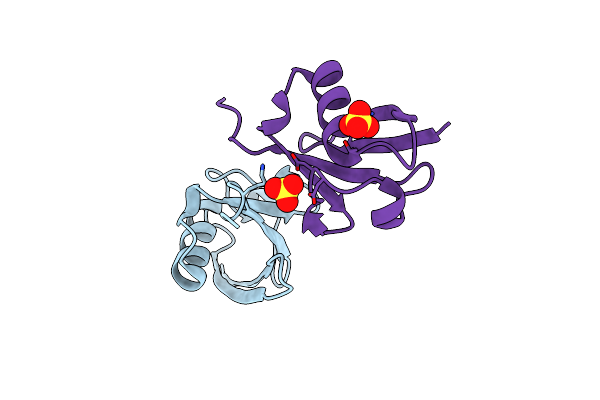
Crystal Structure Of Klebsiella Pneumoniae K1 Capsule-Specific Polysaccharide Lyase In A P1 Crystal Form
Organism: Klebsiella phage ntuh-k2044-k1-1
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2022-05-18 Classification: VIRAL PROTEIN Ligands: LMR, IMD, GOL |
 |
Crystal Structure Of Klebsiella Pneumoniae K1 Capsule-Specific Polysaccharide Lyase In A C2 Crystal Form
Organism: Klebsiella phage ntuh-k2044-k1-1
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2022-05-18 Classification: VIRAL PROTEIN Ligands: CIT, CO3 |

