Search Count: 15
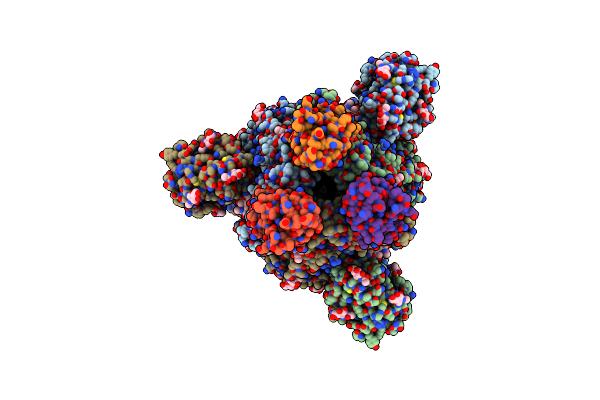 |
Organism: Severe acute respiratory syndrome coronavirus 2, Vicugna pacos
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2021-08-11 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
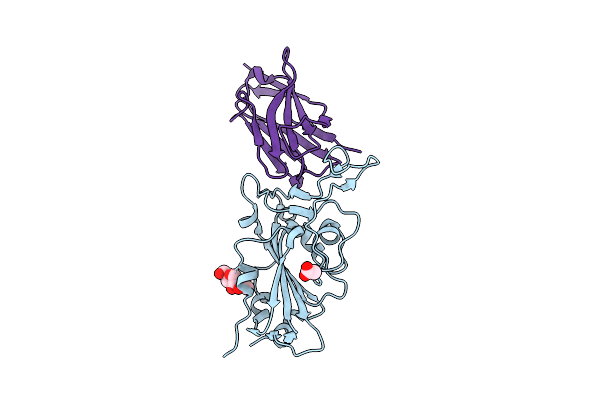 |
Organism: Severe acute respiratory syndrome coronavirus 2, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-08-11 Classification: ANTIVIRAL PROTEIN Ligands: NAG, ACT |
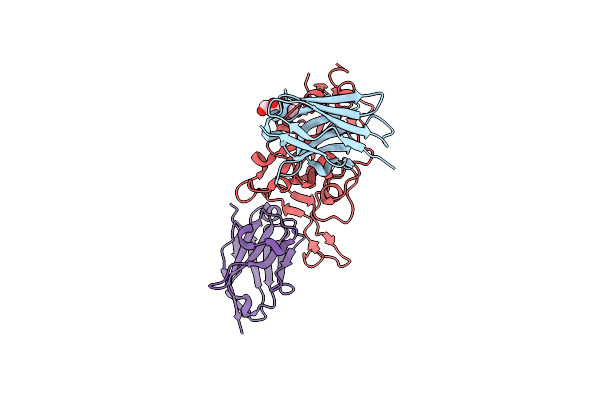 |
Organism: Lama glama, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-08-11 Classification: ANTIVIRAL PROTEIN Ligands: NAG, CIT, CL |
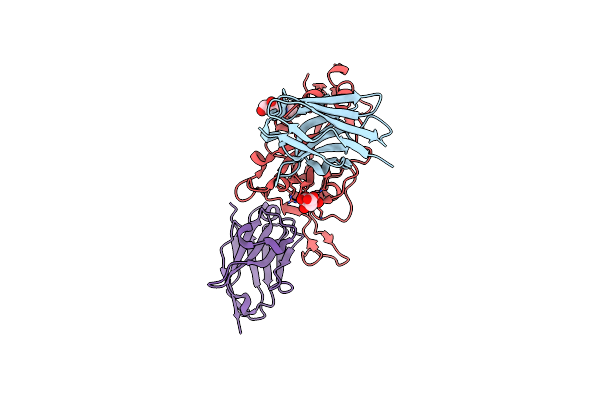 |
Organism: Lama glama, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2021-08-11 Classification: ANTIVIRAL PROTEIN Ligands: NAG, CIT, CL |
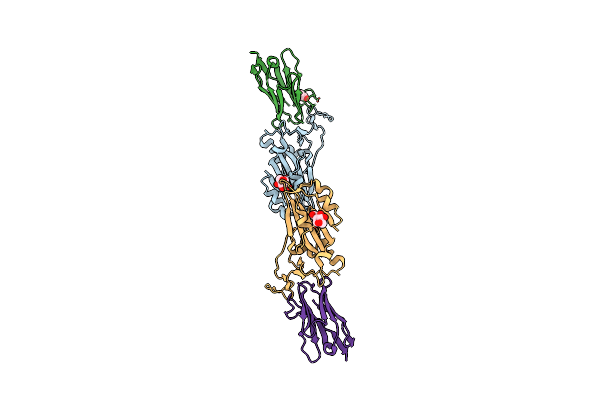 |
Organism: Severe acute respiratory syndrome coronavirus 2, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-08-11 Classification: ANTIVIRAL PROTEIN Ligands: NAG, GOL |
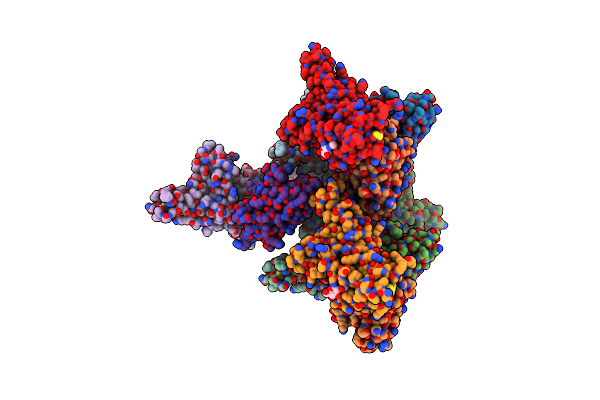 |
Organism: Severe acute respiratory syndrome coronavirus 2, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2021-08-11 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
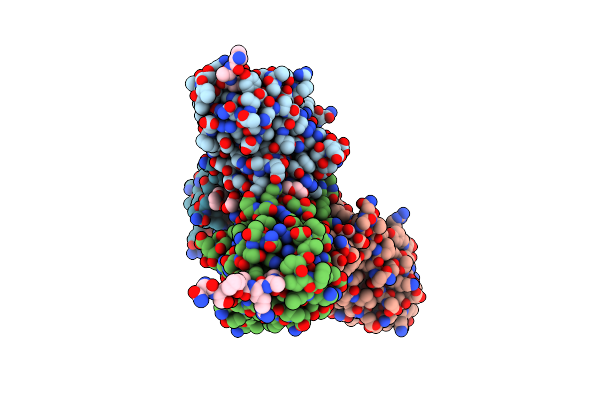 |
Crystal Structure Of Neuropilin-1 B1 Domain In Complex With Sars-Cov-2 S1 C-End Rule (Cendr) Peptide
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2020-10-28 Classification: SIGNALING PROTEIN Ligands: PEG, GOL |
 |
Organism: Hazara virus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-01-13 Classification: VIRAL PROTEIN |
 |
Organism: Human respiratory syncytial virus
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2014-01-22 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Organism: Human respiratory syncytial virus
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2014-01-22 Classification: VIRAL PROTEIN Ligands: CD |
 |
Organism: Human respiratory syncytial virus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-01-22 Classification: VIRAL PROTEIN Ligands: ZN |
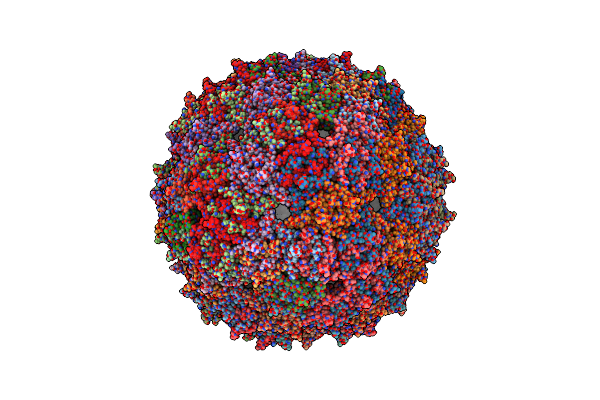 |
Organism: Enterobacteria phage ms2
Method: ELECTRON MICROSCOPY Resolution:39.00 Å Release Date: 2013-07-17 Classification: VIRUS |
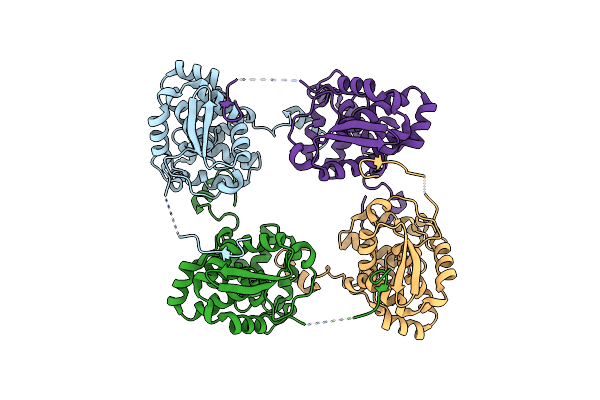 |
Organism: Schmallenberg virus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2013-05-01 Classification: VIRAL PROTEIN |
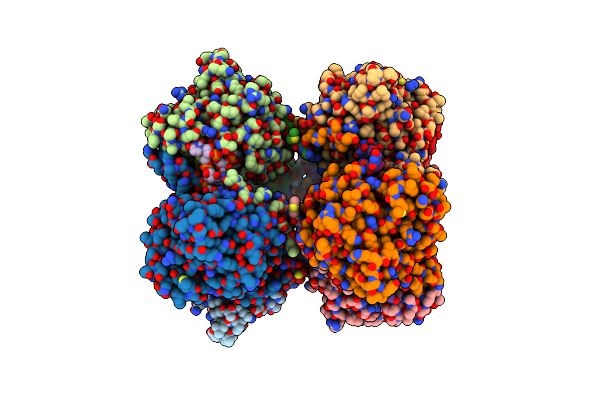 |
Crystal Structure Of The Nucleocapsid Protein From Bunyamwera Virus Bound To Rna
Organism: Bunyamwera virus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2013-05-01 Classification: VIRAL PROTEIN/RNA |
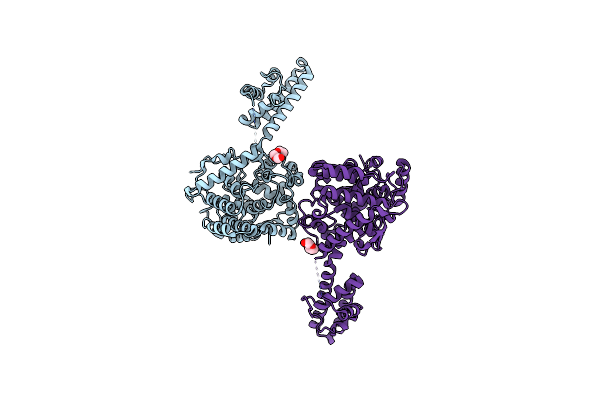 |
Structure Of The Crimean-Congo Haemorrhagic Fever Virus Nucleocapsid Protein
Organism: Crimean-congo hemorrhagic fever virus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-08-22 Classification: VIRAL PROTEIN Ligands: PGE |

