Search Count: 56
 |
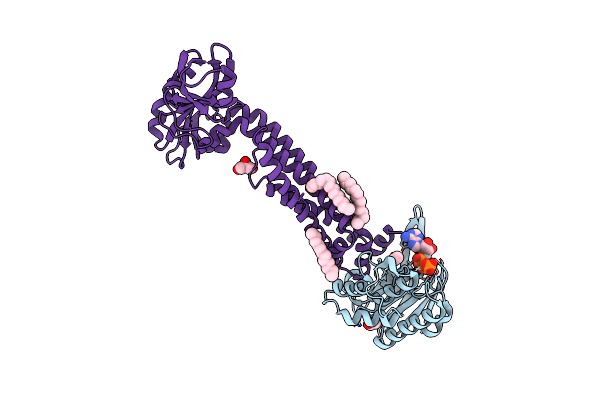
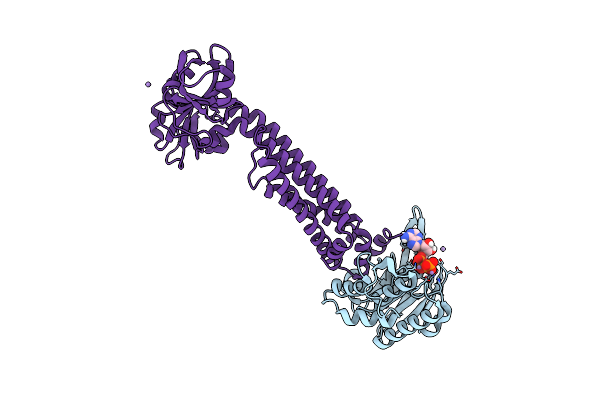
Cryo-Em Structure Of Tmprss2 In Complex With Fab Fragments Of 752 Mab And 2228 Mab
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli, Homo sapiens, Macaca mulatta, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: SIGNALING PROTEIN |
 |
Full Agonist- And Positive Allosteric Modulator-Bound Mu-Type Opioid Receptor-G Protein Complex
Organism: Escherichia coli, Homo sapiens, Macaca mulatta, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: SIGNALING PROTEIN Ligands: VV9 |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab816
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: VIRAL PROTEIN Ligands: NAG |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab803
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: VIRAL PROTEIN Ligands: NAG |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab765
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: VIRAL PROTEIN Ligands: NAG |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab712
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: VIRAL PROTEIN |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab709
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: VIRAL PROTEIN |
 |
The Sars-Cov-2 Receptor Binding Domain Bound With The Fab Fragment Of A Human Neutralizing Antibody Ab847
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: VIRAL PROTEIN |
 |
Organism: Corynebacterium diphtheriae nctc 13129
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN Ligands: MG, ANP, ACT, GOL, MYZ, D12 |
 |
Organism: Corynebacterium diphtheriae nctc 13129
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN Ligands: ATP, MN |
 |
Heme Exporter In Complex With Mn-Containing Protoporphyrin Ix, Mn-Anomalous Data
Organism: Corynebacterium diphtheriae nctc 13129
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN Ligands: AGA, PGE, MNR, PEG, EDO |
 |
Heme Exporter Hrtba In Complex With Protoporphyrin Ix Containing Manganese(Iii), High Resolution Data
Organism: Corynebacterium diphtheriae nctc 13129
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN Ligands: AGA, PGE, MNR, PEG |
 |
Organism: Corynebacterium diphtheriae nctc 13129
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN Ligands: SO4 |
 |
Organism: Corynebacterium diphtheriae nctc 13129
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN Ligands: HEM |
 |
Time-Resolved Serial Femtosecond Crystallography Structure Of Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3: Resting State Structure With Bromide Ion
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: RET, HEX, D10, BR |
 |
Time-Resolved Serial Femtosecond Crystallography Structure Of Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3 : Structure Obtained 1 Msec After Photoexcitation With Bromide Ion
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: RET, HEX, D10, BR |
 |
Anion Free Form Of Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3, Structure Determined By Serial Femtosecond Crystallography At Sacla
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-02-16 Classification: MEMBRANE PROTEIN Ligands: RET, HEX, DD9, C14, R16, OCT, CL |
 |
Structure Of Reaction Intermediate Of Cytochrome P450 No Reductase (P450Nor) Determined By Xfel
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-05-19 Classification: OXIDOREDUCTASE Ligands: HEM, NO, GOL |
 |
Organism: Persicobacter sp. ccb-qb2
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-12-30 Classification: SUGAR BINDING PROTEIN Ligands: CA |

