Search Count: 36
 |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2025-04-30 Classification: HYDROLASE Ligands: OCA, PPI, GOL, A1L60, FMT, ZN, CA, 6NA, CL, BUA, 11A |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2024-12-25 Classification: IMMUNE SYSTEM/DE NOVO PROTEIN Ligands: SO4 |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-12-11 Classification: IMMUNE SYSTEM/DE NOVO PROTEIN |
 |
 |
 |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: TRANSLOCASE |
 |
Crystal Structure Of Aedes Aegypti Noppera-Bo, Glutathione S-Transferase Epsilon 8, In Glutathione-Bound Form
Organism: Aedes aegypti
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2022-01-26 Classification: TRANSFERASE Ligands: GSH, CA |
 |
Crystal Structure Of Aedes Aegypti Noppera-Bo, Glutathione S-Transferase Epsilon 8, In Daidzein- And Glutathione-Bound Form
Organism: Aedes aegypti
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-01-26 Classification: TRANSFERASE Ligands: GSH, ZF1, CA |
 |
Crystal Structure Of Aedes Aegypti Noppera-Bo, Glutathione S-Transferase Epsilon 8, In Luteolin- And Glutathione-Bound Form
Organism: Aedes aegypti
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-01-26 Classification: TRANSFERASE Ligands: GSH, LU2, CA |
 |
Crystal Structure Of Aedes Aegypti Noppera-Bo, Glutathione S-Transferase Epsilon 8, In Desmethylglycitein And Glutathione-Bound Form
Organism: Aedes aegypti
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2022-01-26 Classification: TRANSFERASE Ligands: GSH, 674, CA |
 |
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Tdp013-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: H1X, DMS, CL |
 |
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Tdp013-, And Gsh-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: H1X, GSH, DTT |
 |
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Tdp015- And Gsh-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: GSH, H29 |
 |
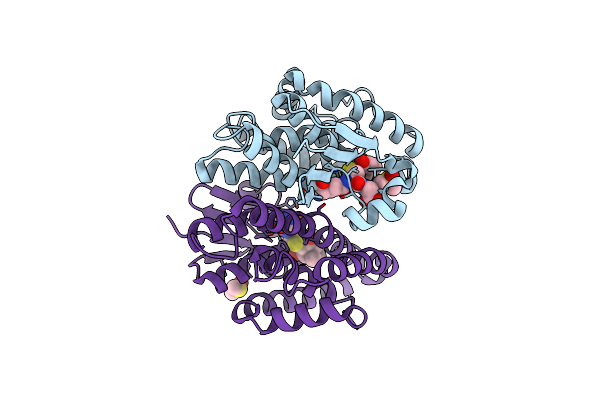
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Dimedone-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: DC1, DMS |
 |
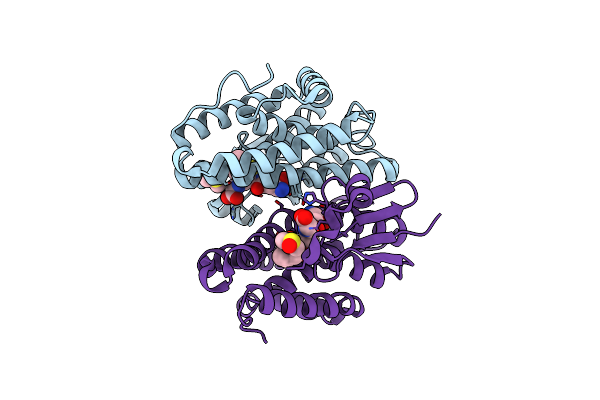
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Dimedone- And Glutathione-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: DC1, GSH, DMS |
 |
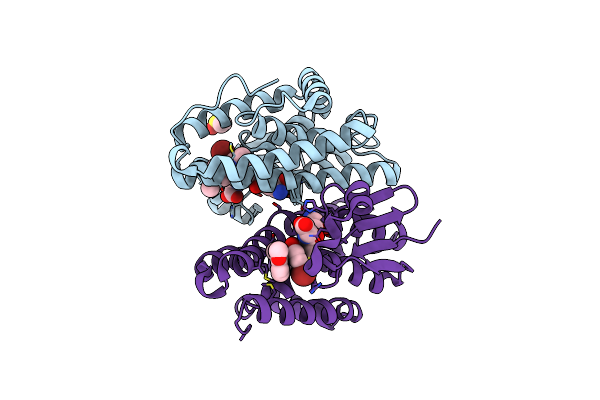
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Gs-Dimedone-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: DC1, GSH, DMS |
 |
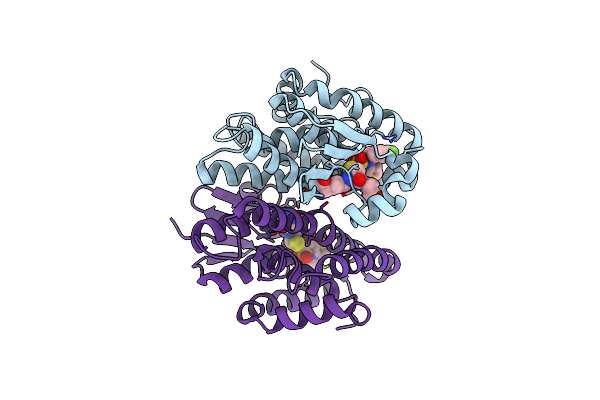
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Tdp011-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: H2X, GSH, DMS |
 |
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Tdp012- And Glutathione-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: H2C, GSH |

