Search Count: 19
 |
Crystal Structure Of Escherichia Coli Gamma-Glutamyltranspeptidase In Complex With Peptidyl Phosphonate Inhibitor 1B
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-09-28 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 6FY, CA |
 |
Crystal Structure Of An Inward-Facing Eukaryotic Abc Multidrug Transporter G277V/A278V/A279V Mutant In Complex With An Cyclic Peptide Inhibitor, Acap
Organism: Cyanidioschyzon merolae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-04-30 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: DMU, TRS |
 |
Organism: Camellia sinensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-04-23 Classification: HYDROLASE Ligands: NAG |
 |
Beta-Primeverosidase In Complex With Disaccharide Substrate-Analog N-Beta-Primeverosylamidine, Natural Aglycone Derivative
Organism: Camellia sinensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-04-23 Classification: HYDROLASE Ligands: VPA, NAG |
 |
Beta-Primeverosidase In Complex With Disaccharide Substrate-Analog N-Beta-Primeverosylamidine, Artificial Aglycone Derivative
Organism: Camellia sinensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-04-23 Classification: HYDROLASE Ligands: ZPA, NAG |
 |
Organism: Cyanidioschyzon merolae
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2014-03-19 Classification: TRANSPORT PROTEIN Ligands: DMU |
 |
Crystal Structure Of An Inward-Facing Eukaryotic Abc Multitrug Transporter G277V/A278V/A279V Mutant
Organism: Cyanidioschyzon merolae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-03-19 Classification: TRANSPORT PROTEIN Ligands: DMU |
 |
Crystal Structure Of Gamma-Glutamyltranspeptidase From Bacillus Subtilis (Crystal Soaked For 0 Min. In Acivicin Soln. )
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-02-19 Classification: HYDROLASE, TRANSFERASE |
 |
Crystal Structure Of Gamma-Glutamyltranspeptidase From Bacillus Subtilis (Crystal Soaked For 3Min. In Acivicin Soln. )
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2014-02-19 Classification: HYDROLASE, TRANSFERASE |
 |
Crystal Structure Of Bacillus Subtilis Gamma-Glutamyltranspeptidase In Complex With Acivicin
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-02-19 Classification: HYDROLASE, TRANSFERASE Ligands: AVN |
 |
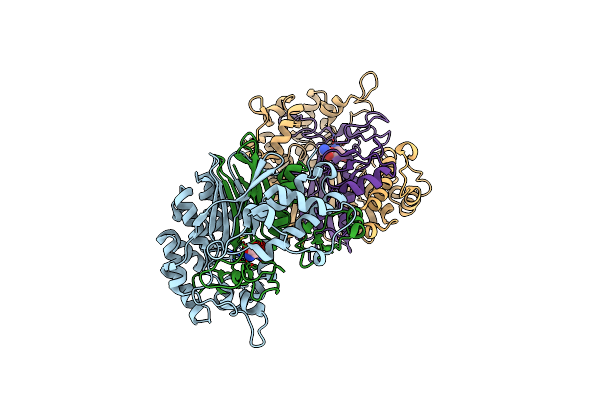
Crystal Structure Of Escherichia Coli Gamma-Glutamyltranspeptidase In Complex With Azaserine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2008-06-24 Classification: TRANSFERASE Ligands: AZS |
 |
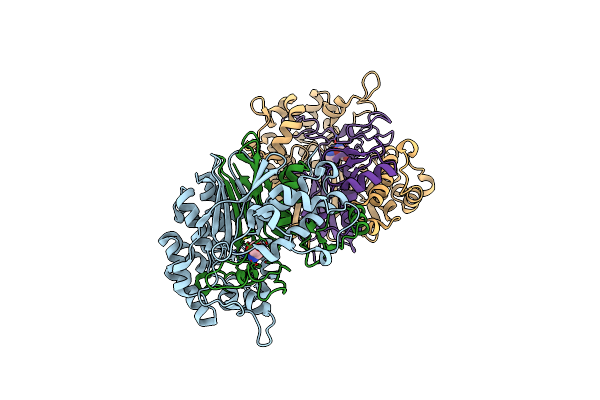
Crystal Structure Of Escherichia Coli Gamma-Glutamyltranspeptidase In Complex With Azaserine Prepared In The Dark
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2008-06-24 Classification: TRANSFERASE Ligands: AZS |
 |
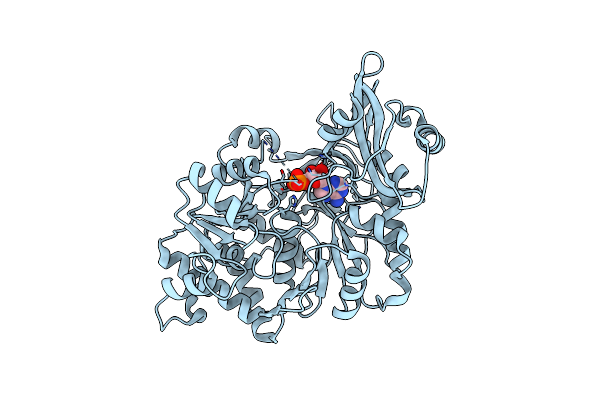
Crystal Structure Of Escherichia Coli Gamma-Glutamyltranspeptidase In Complex With Acivicin
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2008-06-24 Classification: TRANSFERASE Ligands: AVN |
 |
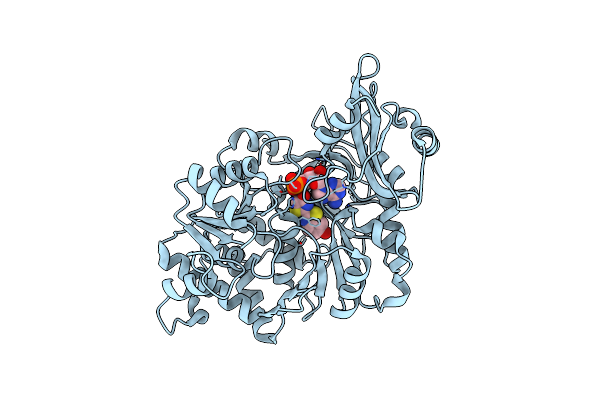
Crystal Structure Of The Thermostable Japanese Firefly Luciferase Complexed With Mgatp
Organism: Luciola cruciata
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-03-21 Classification: OXIDOREDUCTASE Ligands: AMP |
 |
Crystal Structure Of The Thermostable Japanese Firefly Luciferase Complexed With Oxyluciferin And Amp
Organism: Luciola cruciata
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2006-03-21 Classification: OXIDOREDUCTASE Ligands: AMP, OLU |
 |
Crystal Structure Of The Thermostable Japanese Firefly Luciferase Complexed With High-Energy Intermediate Analogue
Organism: Luciola cruciata
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2006-03-21 Classification: OXIDOREDUCTASE Ligands: CL, SLU |
 |
Crystal Structure Of The Thermostable Japanese Firefly Luciferase Red-Color Emission S286N Mutant Complexed With High-Energy Intermediate Analogue
Organism: Luciola cruciata
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2006-03-21 Classification: OXIDOREDUCTASE Ligands: CL, SLU |
 |
Crystal Structure Of Gamma-Glutamylcysteine Synthetase From Escherichia Coli B
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2004-10-05 Classification: LIGASE |
 |
Crystal Structure Of Gamma-Glutamylcysteine Synthetase From Escherichia Coli B Complexed With Transition-State Analogue
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-10-05 Classification: LIGASE Ligands: MG, P2S, ADP, P6G |

