Search Count: 60
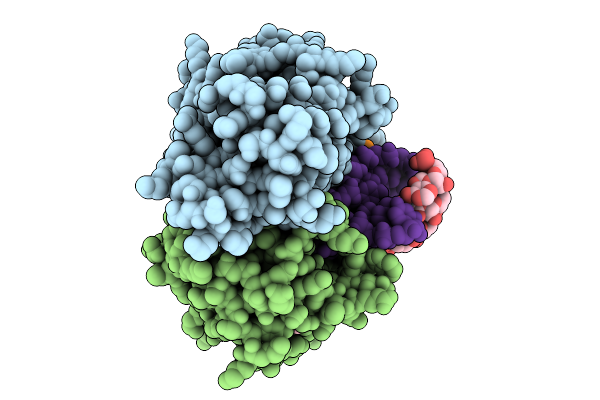 |
Structural Basis Of Substrate Promiscuity In The Archaeal Rna-Splicing Endonuclease From Candidatus Micrarchaeum Acidiphilum (Arman-2)
Organism: Candidatus micrarchaeum acidiphilum arman-2, Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: SPLICING/RNA Ligands: ACY, A23 |
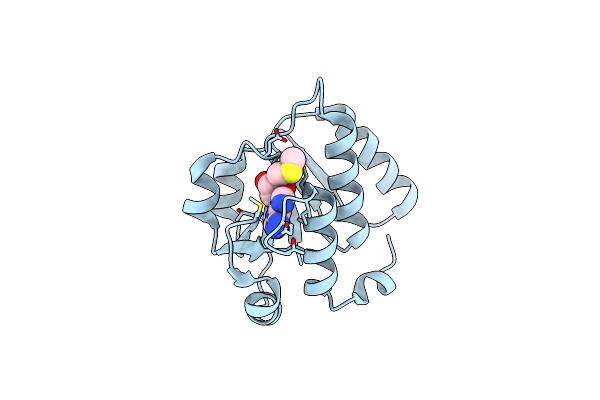 |
The X-Ray Structure Of N-Terminal Catalytic Domain Of Thermoplasma Acidophilum Trna Methyltransferase Trm56 (Ta0931) In Complex With 5'-Methylthioadenosine
Organism: Thermoplasma acidophilum dsm 1728
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSFERASE Ligands: MTA |
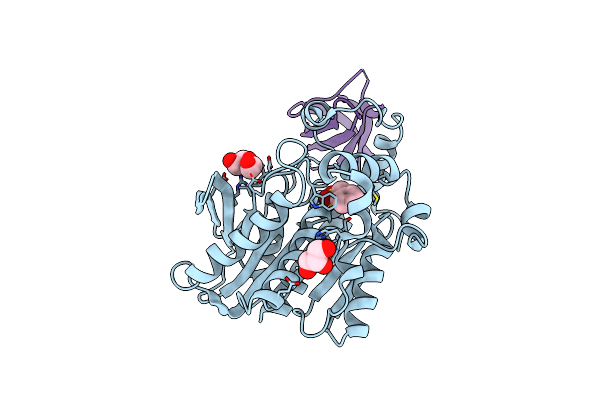 |
Crystal Structure Of The Thermotoga Maritima Cold Shock Protein Mutant Est#13 In Complex With Aacest
Organism: Alicyclobacillus acidocaldarius, Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-15 Classification: PROTEIN BINDING Ligands: PMS, PEG, GOL |
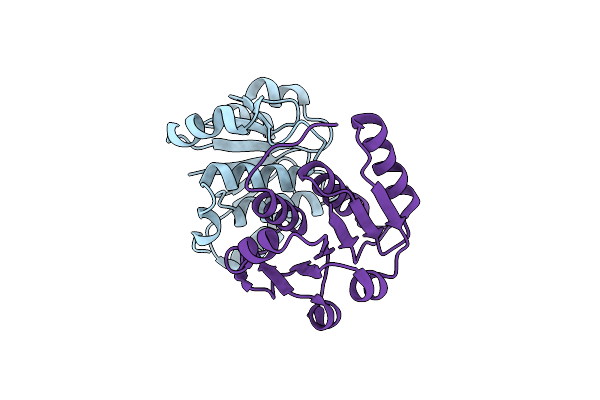 |
The X-Ray Structure Of N-Terminal Catalytic Domain Of Thermoplasma Acidophilum Trna Methyltransferase Trm56 (Ta0931).
Organism: Thermoplasma acidophilum dsm 1728
Method: X-RAY DIFFRACTION Release Date: 2024-11-27 Classification: TRANSFERASE |
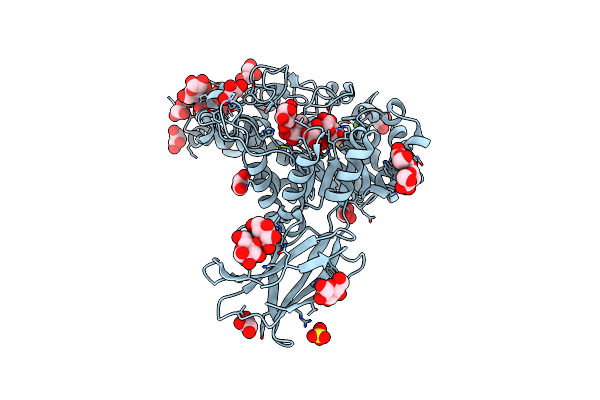 |
The 1.26 Angstrom Resolution Structure Of Bacillus Cereus Beta-Amylase In Complex With Maltose
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2024-09-25 Classification: HYDROLASE Ligands: CA, SO4, GOL, BGC |
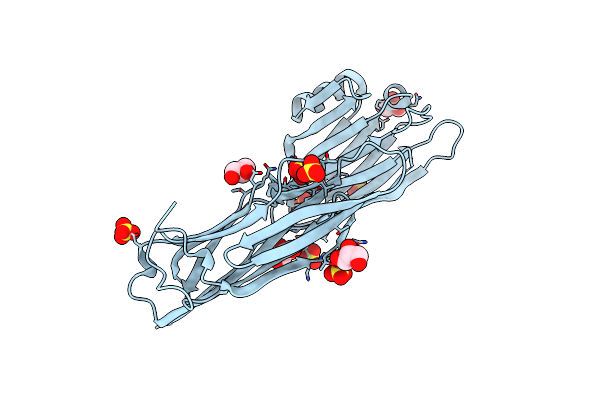 |
Crystal Structure Of The Collagen Binding Domain Of Cnm From Streptococcus Mutans
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-04-10 Classification: PROTEIN BINDING Ligands: SO4, GOL |
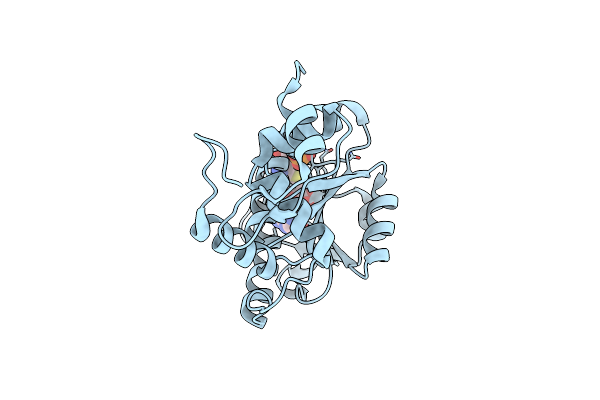 |
Crystal Structure Of Mutant Trna [Gm18] Methyltransferase Trmh (E107G) In Complex With S-Adenosyl-L-Methionine From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-03-23 Classification: TRANSFERASE Ligands: SAM, PO4 |
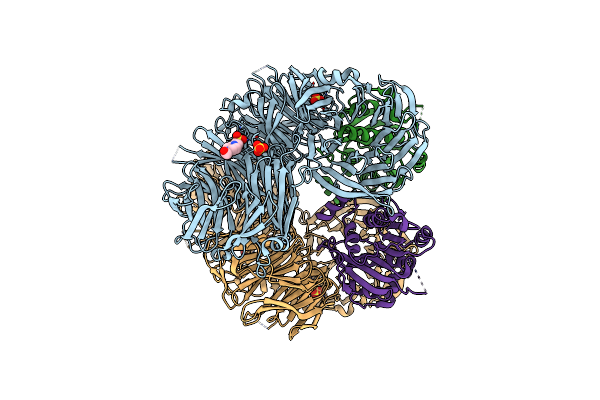 |
The X-Ray Structure Of Yeast Trna Methyltransferase Complex Of Trm7 And Trm734 Essential For 2'-O-Methylation At The First Position Of Anticodon In Specific Trnas
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-10-02 Classification: TRANSFERASE Ligands: SO4, EPE |
 |
The X-Ray Structure Of Yeast Trna Methyltransferase Trm7-Trm734 In Complex With S-Adenosyl-L-Methionine
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2019-10-02 Classification: TRANSFERASE Ligands: EPE, SO4, SAM |
 |
The X-Ray Crystal Structure Of Subunit Fusion Rna Splicing Endonuclease From Methanopyrus Kandleri
Organism: Methanopyrus kandleri av19
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2018-01-24 Classification: HYDROLASE Ligands: PO4 |
 |
The X-Ray Crystal Structure Of A Tetr Family Transcription Regulator Rcda Involved In The Regulation Of Biofilm Formation In Escherichia Coli
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2017-12-27 Classification: TRANSCRIPTION |
 |
Crystal Structure Of The Archaeal Trna M2G/M22G10 Methyltransferase (Atrm11) From Thermococcus Kodakarensis
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-07-06 Classification: TRANSFERASE |
 |
Crystal Structure Of The Archaeal Trna M2G/M22G10 Methyltransferase (Atrm11) In Complex With S-Adenosyl-L-Methionine (Sam) From Thermococcus Kodakarensis
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2016-07-06 Classification: TRANSFERASE Ligands: SAM |
 |
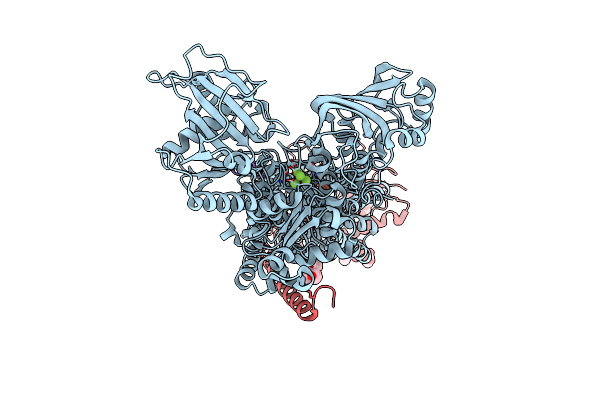
Kinetics By X-Ray Crystallography: Tl+-Substitution Of Bound K+ In The E2.Mgf42-.2K+ Crystal After 0.75 Min.
Organism: Squalus acanthias
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-09-02 Classification: HYDROLASE/TRANSPORT PROTEIN Ligands: MF4, MG, K, TL, CLR, NAG |
 |
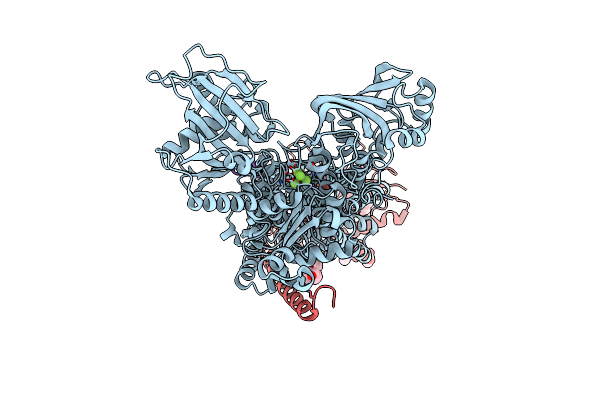
Kinetics By X-Ray Crystallography: Tl+-Substitution Of Bound K+ In The E2.Mgf42-.2K+ Crystal After 1.5 Min
Organism: Squalus acanthias
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-09-02 Classification: HYDROLASE/TRANSPORT PROTEIN Ligands: MF4, MG, K, TL, CLR, NAG |
 |
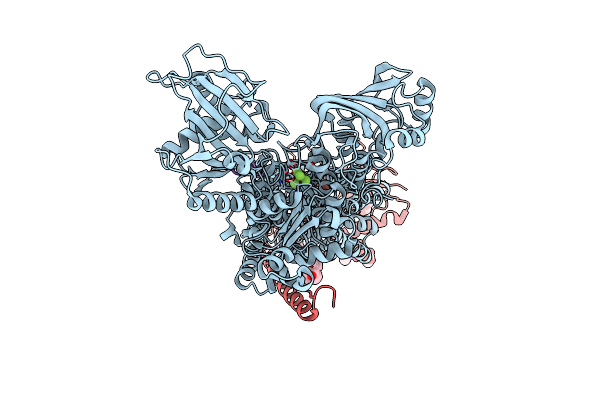
Kinetics By X-Ray Crystallography: Tl+-Substitution Of Bound K+ In The E2.Mgf42-.2K+ Crystal After 3.5 Min
Organism: Squalus acanthias
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-09-02 Classification: HYDROLASE/TRANSPORT PROTEIN Ligands: MF4, MG, K, TL, CLR, NAG |
 |
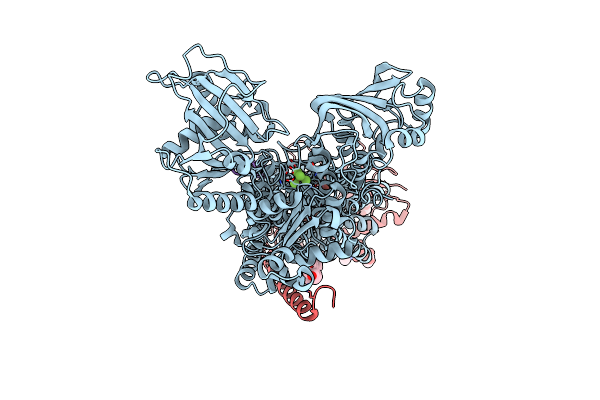
Kinetics By X-Ray Crystallography: Tl+-Substitution Of Bound K+ In The E2.Mgf42-.2K+ Crystal After 5 Min
Organism: Squalus acanthias
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-09-02 Classification: HYDROLASE/TRANSPORT PROTEIN Ligands: MF4, MG, K, TL, CLR, NAG |
 |
Kinetics By X-Ray Crystallography: Tl+-Substitution Of Bound K+ In The E2.Mgf42-.2K+ Crystal After 7.0 Min
Organism: Squalus acanthias
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2015-09-02 Classification: HYDROLASE/TRANSPORT PROTEIN Ligands: MF4, MG, K, TL, CLR, NAG |
 |
Kinetics By X-Ray Crystallography: Tl+-Substitution Of Bound K+ In The E2.Mgf42-.2K+ Crystal After 8.5 Min
Organism: Squalus acanthias
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-09-02 Classification: HYDROLASE/TRANSPORT PROTEIN Ligands: MF4, MG, TL, K, CLR, NAG |
 |
Kinetics By X-Ray Crystallography: Tl+-Substitution Of Bound K+ In The E2.Mgf42-.2K+ Crystal After 16.5 Min
Organism: Squalus acanthias
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-09-02 Classification: HYDROLASE/TRANSPORT PROTEIN Ligands: MF4, MG, TL, K, CLR, NAG |

