Search Count: 13
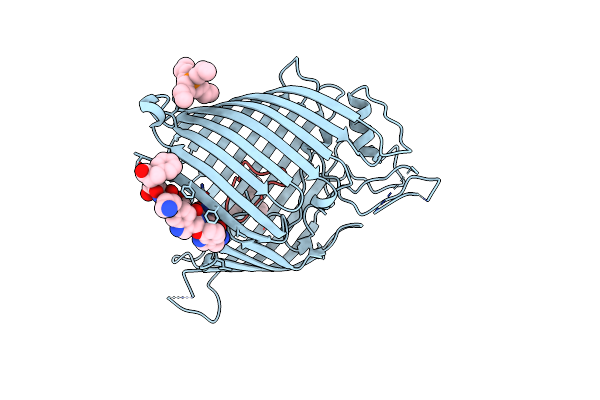 |
Organism: Escherichia coli, Synthetic construct, Photorhabdus khanii
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN Ligands: 4NE |
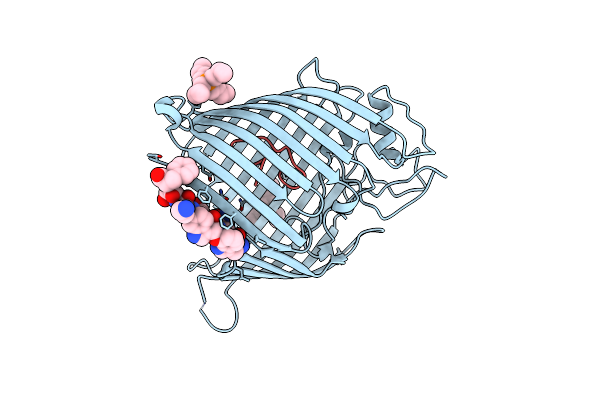 |
Organism: Escherichia coli, Synthetic construct, Photorhabdus khanii
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN Ligands: 4NE |
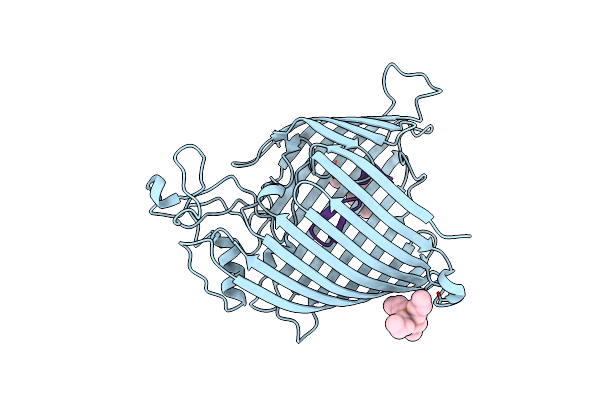 |
Organism: Escherichia coli o139:h28 (strain e24377a / etec), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN Ligands: 4NE |
 |
Crystal Structure Of An Inward-Facing Eukaryotic Abc Multidrug Transporter G277V/A278V/A279V Mutant In Complex With An Cyclic Peptide Inhibitor, Acap
Organism: Cyanidioschyzon merolae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-04-30 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: DMU, TRS |
 |
Organism: Cyanidioschyzon merolae
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2014-03-19 Classification: TRANSPORT PROTEIN Ligands: DMU |
 |
Crystal Structure Of An Inward-Facing Eukaryotic Abc Multitrug Transporter G277V/A278V/A279V Mutant
Organism: Cyanidioschyzon merolae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-03-19 Classification: TRANSPORT PROTEIN Ligands: DMU |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2013-06-12 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: OLC |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2013-04-03 Classification: TRANSPORT PROTEIN Ligands: OLC |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-04-03 Classification: TRANSPORT PROTEIN Ligands: OLC |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2013-04-03 Classification: TRANSPORT PROTEIN Ligands: BNU |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2013-04-03 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: OLC |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-04-03 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: OLC |
 |
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-04-03 Classification: TRANSPORT PROTEIN Ligands: OLC |

