Search Count: 18
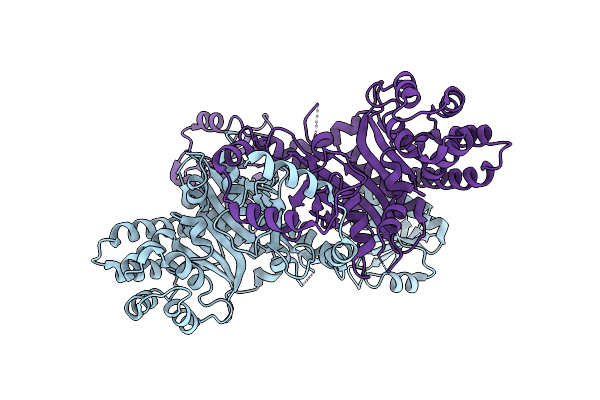 |
Organism: Thermochaetoides thermophila
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-08-21 Classification: TRANSFERASE |
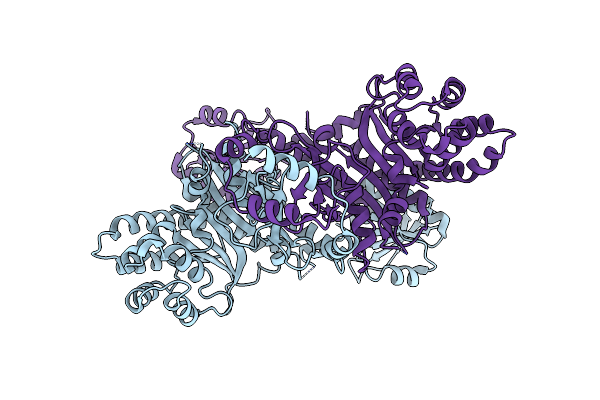 |
Crystal Structure Of Chaetomium Thermophilum Gcn2 Hisrs-Like Domain, F1090C/G1326S Variant
Organism: Thermochaetoides thermophila
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2024-08-21 Classification: TRANSFERASE |
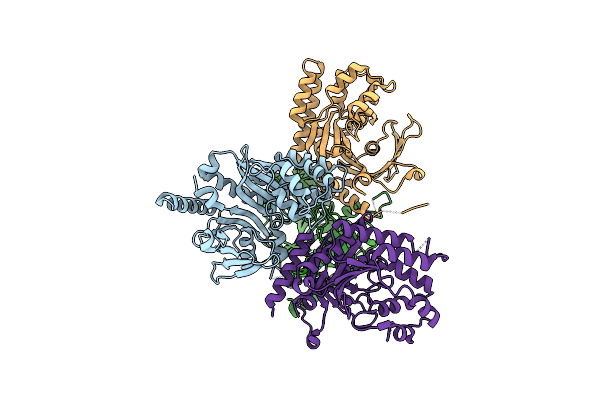 |
Crystal Structure Of Chaetomium Thermophilum Gcn2 Hisrs-Like Domain, Catalytic Domain
Organism: Thermochaetoides thermophila
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2024-08-21 Classification: TRANSLATION |
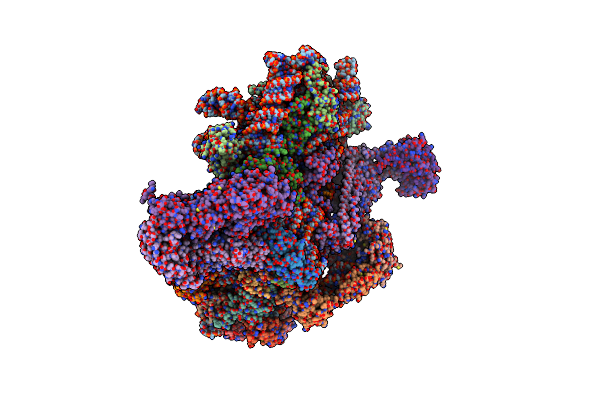 |
Structure Of A Partial Yeast 48S Preinitiation Complex In Open Conformation.
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Resolution:5.15 Å Release Date: 2019-07-31 Classification: RIBOSOME Ligands: 7NO, MG, ZN, GCP |
 |
Structure Of A Partial Yeast 48S Preinitiation Complex In Closed Conformation
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Kluyveromyces lactis (strain atcc 8585 / cbs 2359 / dsm 70799 / nbrc 1267 / nrrl y-1140 / wm37)
Method: ELECTRON MICROSCOPY Release Date: 2019-06-26 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Structure Of A Partial Yeast 48S Preinitiation Complex With Eif5 N-Terminal Domain (Model C1)
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae, Kluyveromyces lactis (strain atcc 8585 / cbs 2359 / dsm 70799 / nbrc 1267 / nrrl y-1140 / wm37)
Method: ELECTRON MICROSCOPY Release Date: 2018-12-05 Classification: RIBOSOME Ligands: MG, ZN, MET, GCP |
 |
Structure Of A Partial Yeast 48S Preinitiation Complex With Eif5 N-Terminal Domain (Model C2)
Organism: Saccharomyces cerevisiae, Kluyveromyces lactis (strain atcc 8585 / cbs 2359 / dsm 70799 / nbrc 1267 / nrrl y-1140 / wm37)
Method: ELECTRON MICROSCOPY Resolution:3.02 Å Release Date: 2018-12-05 Classification: RIBOSOME Ligands: MG, ZN, MET, GCP |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: SOLUTION NMR Release Date: 2017-05-31 Classification: TRANSLATION |
 |
Organism: Saccharomyces cerevisiae s288c
Method: SOLUTION NMR Release Date: 2016-10-26 Classification: TRANSLATION |
 |
Organism: Saccharomyces cerevisiae, Kluyveromyces lactis
Method: ELECTRON MICROSCOPY Release Date: 2015-08-12 Classification: TRANSLATION Ligands: MG, ZN |
 |
Structure Of A Partial Yeast 48S Preinitiation Complex In Closed Conformation
Organism: Saccharomyces cerevisiae, Kluyveromyces lactis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2015-08-12 Classification: TRANSLATION Ligands: MG, ZN, MET, GCP |
 |
Organism: Saccharomyces cerevisiae, Kluyveromyces lactis
Method: ELECTRON MICROSCOPY Release Date: 2014-11-05 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Saccharomyces cerevisiae, Synthetic construct, Kluyveromyces lactis
Method: ELECTRON MICROSCOPY Release Date: 2014-11-05 Classification: RIBOSOME Ligands: MG, ZN, MET |
 |
Crystal Structure Of Eif2Alpha Protein Kinase Gcn2: D835N Inactivating Mutant In Apo Form
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2005-06-21 Classification: TRANSFERASE Ligands: GOL |
 |
Crystal Structure Of Eif2Alpha Protein Kinase Gcn2: R794G Hyperactivating Mutant In Apo Form.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2005-06-21 Classification: TRANSFERASE Ligands: GOL |
 |
Crystal Structure Of Eif2Alpha Protein Kinase Gcn2: R794G Hyperactivating Mutant Complexed With Amppnp.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2005-06-21 Classification: TRANSFERASE Ligands: MG, ANP |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2005-06-21 Classification: TRANSFERASE |
 |
Crystal Structure Of Eif2Alpha Protein Kinase Gcn2: Wild-Type Complexed With Atp.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2005-06-21 Classification: TRANSFERASE Ligands: MG, ATP |

