Search Count: 53
 |
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-05-04 Classification: HYDROLASE |
 |
Crystal Structure Of The Gh12 Domain From Acidothermus Cellulolyticus Guxa Bound To Cellobiose
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-05-04 Classification: HYDROLASE Ligands: ZN, CL, NA, ACT, GOL |
 |
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-12-23 Classification: OXIDOREDUCTASE Ligands: NAD, GOL, EDO, NA, ADP |
 |
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-12-23 Classification: OXIDOREDUCTASE Ligands: NAD, HBR, HBS, ADP |
 |
Structure Of Serratia Marcescens 2,3-Butanediol Dehydrogenase Mutant Q247A/V139Q
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-12-23 Classification: OXIDOREDUCTASE Ligands: NAD, NA, EDO |
 |
Structure Of Caldicellulosiruptor Danielii Cbm3 Module Of Glycoside Hydrolase Wp_045175321
Organism: Caldicellulosiruptor sp. wai35.b1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-04-24 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Structure Of Caldicellulosiruptor Danielii Gh10 Module Of Glycoside Hydrolase Wp_045175321
Organism: Caldicellulosiruptor sp. wai35.b1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-04-24 Classification: HYDROLASE Ligands: SO4, EDO, GOL, FMT |
 |
Structure Of Caldicellulosiruptor Danielii Gh48 Module Of Glycoside Hydrolase Wp_045175321
Organism: Caldicellulosiruptor sp. wai35.b1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-04-24 Classification: HYDROLASE Ligands: CA |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-02-20 Classification: TRANSFERASE Ligands: CO, NAG, CA |
 |
The Crystal Structure Of Caldicellulosiruptor Kristjanssonii Tapirin C-Terminal Domain
Organism: Caldicellulosiruptor kristjanssonii (strain atcc 700853 / dsm 12137 / i77r1b)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2018-12-19 Classification: CELL ADHESION Ligands: CA, GOL |
 |
The Crystal Structure Of Caldicellulosiruptor Hydrothermalis Tapirin C-Terminal Domain
Organism: Caldicellulosiruptor hydrothermalis (strain dsm 18901 / vkm b-2411 / 108)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-12-19 Classification: CELL ADHESION Ligands: ZN, EDO, GOL, CL, GOA, OXL |
 |
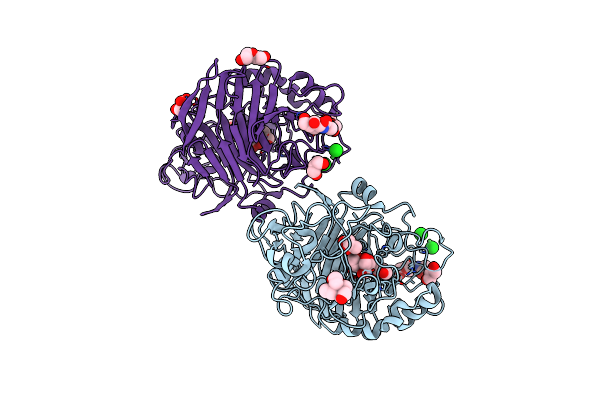
Organism: Hypocrea atroviridis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-01-31 Classification: HYDROLASE Ligands: NAG, NI, GOL, BTB, GS1, GLC, PEG |
 |
Organism: Hypocrea atroviridis (strain atcc 20476 / imi 206040)
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2018-01-31 Classification: HYDROLASE Ligands: NAG, NI, BTB, GOL, CL, PEG |
 |
Organism: Zymomonas mobilis
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2017-10-18 Classification: CELL ADHESION Ligands: TPP, MG, SO4, EDO |
 |
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-02-01 Classification: LYASE Ligands: CU, GOL, IOD |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2016-09-28 Classification: CELL ADHESION Ligands: MES, CL, K, EDO, GOL |
 |
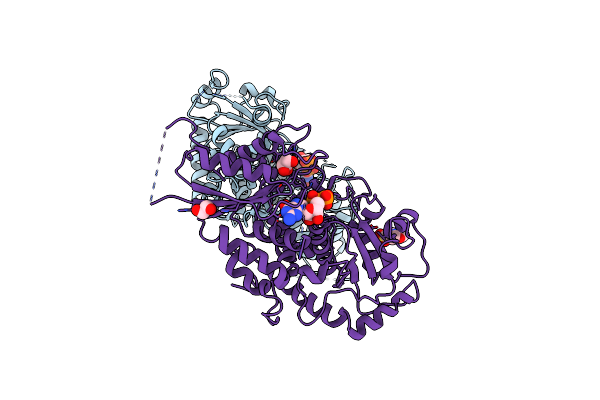
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-09-28 Classification: CELL ADHESION Ligands: GDP, MES, EDO, GOL |
 |
The Structure Of Arabidopsis Thaliana Fut1 Mutant R284K In Complex With Gdp
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-09-28 Classification: CELL ADHESION Ligands: GDP, GOL, EDO, MES |
 |
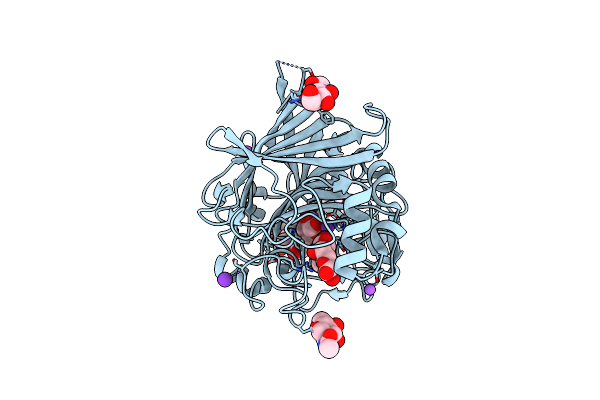
The Structure Of Bacillus Pumilus Gh48 In Complex With Cellobiose And Cellohexaose
Organism: Bacillus pumilus (strain safr-032)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-08-10 Classification: HYDROLASE Ligands: CA, MLI, EDO, GOL, NA |
 |
Organism: Penicillium funiculosum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-06-22 Classification: HYDROLASE Ligands: NAG, K, NA, CL |

