Search Count: 24
 |
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: CELL ADHESION |
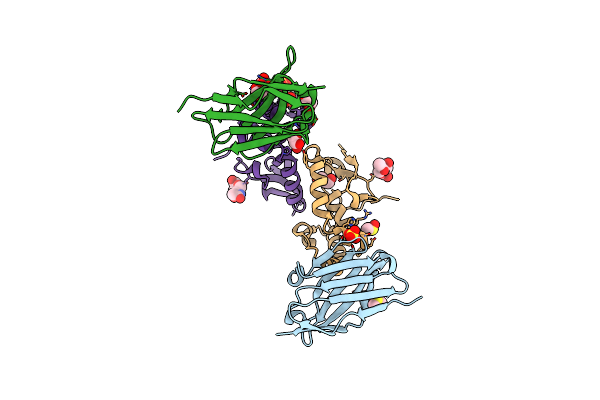 |
Crystal Structure Of The Fzd3 Cysteine-Rich Domain In Complex With A Nanobody (14478)
Organism: Lama glama, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-08-28 Classification: SIGNALING PROTEIN Ligands: DMS, GOL, CIT, NAG, SO4 |
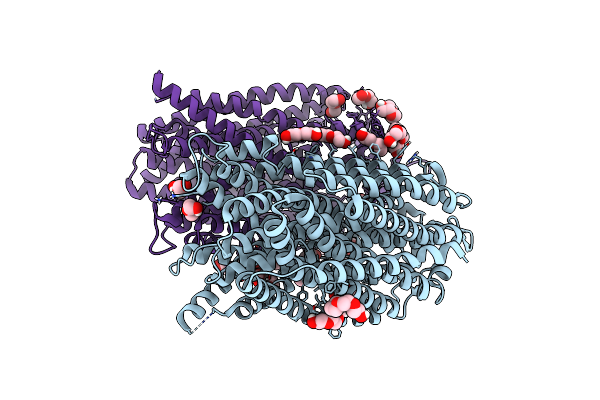 |
Time-Resolved Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima 0-60-Seconds Post Reaction Initiation With Na+
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: LMT, PEG, PGE, PG4 |
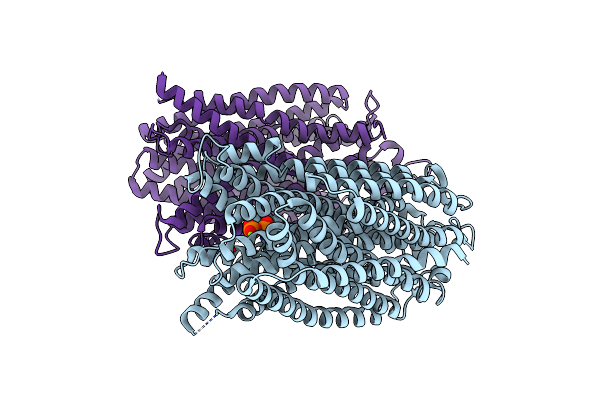 |
Time-Resolved Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima 300-Seconds Post Reaction Initiation With Na+
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:3.98 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: DPO, MG |
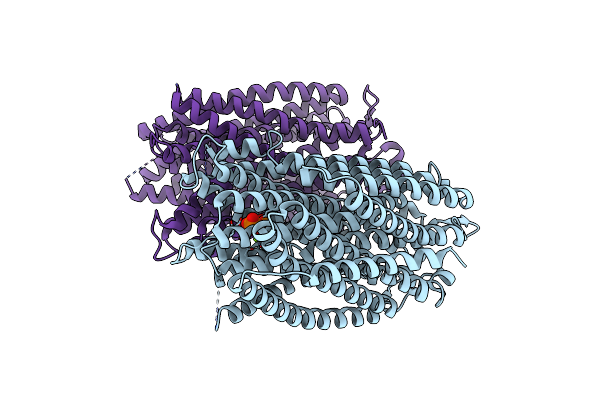 |
Time-Resolved Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima 600-Seconds Post Reaction Initiation With Na+
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:3.84 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: DPO, MG |
 |
Time-Resolved Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima 3600-Seconds Post Reaction Initiation With Na+
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:4.53 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: DPO, MG, K, PO4 |
 |
Crystal Structure Of Pyrobaculum Aerophilum Potassium-Independent Proton Pumping Membrane Integral Pyrophosphatase In Complex With Imidodiphosphate And Magnesium, And With Bound Sulphate
Organism: Pyrobaculum aerophilum
Method: X-RAY DIFFRACTION Resolution:3.84 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: MG, 2PN, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2020-11-11 Classification: SIGNALING PROTEIN Ligands: NAG, SO4, EDO, B1J |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-11-11 Classification: SIGNALING PROTEIN Ligands: NAG, SO4, DMS, EDO, QR2 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2020-10-28 Classification: SIGNALING PROTEIN Ligands: SO4, DMS, EDO, NAG, TEP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2020-10-28 Classification: SIGNALING PROTEIN Ligands: SO4, NAG, DMS, CFF, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2020-09-16 Classification: HYDROLASE Ligands: NAG, SO4, DMS, EDO, PJK |
 |
Structure Of Notum In Complex With A 1-(3-Chlorophenyl)-2,5-Dimethyl-1H-Pyrrole-3-Carboxylic Acid Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2020-08-19 Classification: HYDROLASE Ligands: PZ8, NAG, DMS, SO4, EDO |
 |
Structure Of The Wnt Deacylase Notum In Complex With A Pyrrolidine-3-Carboxylic Acid Fragment 587
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-05-13 Classification: HYDROLASE Ligands: SO4, EDO, PQT, NAG |
 |
Structure Of The Wnt Deacylase Notum In Complex With A Pyrrolidine-3-Carboxylic Acid Fragment 598
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-05-13 Classification: HYDROLASE Ligands: NAG, PUE, SO4, EDO |
 |
Structure Of The Wnt Deacylase Notum In Complex With A Pyrrole-3-Carboxylic Acid Fragment 454
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2020-05-06 Classification: HYDROLASE Ligands: SO4, PQZ, NAG |
 |
Structure Of The Wnt Deacylase Notum In Complex With A Pyrrole-3-Carboxylic Acid Fragment 471
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-05-06 Classification: HYDROLASE Ligands: SO4, DMS, PQH, NAG |
 |
Structure Of The Wnt Deacylase Notum In Complex With A Pyrrole-3-Carboxylic Acid Fragment 686
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-05-06 Classification: HYDROLASE Ligands: PQK, NAG, SO4, EDO |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2020-03-25 Classification: SIGNALING PROTEIN Ligands: NAG, SO4 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2020-03-04 Classification: ONCOPROTEIN |

