Search Count: 17
 |
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2023-05-24 Classification: HYDROLASE Ligands: TT6, GOL |
 |
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2023-05-24 Classification: HYDROLASE Ligands: GOL, TU5 |
 |
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2023-05-24 Classification: HYDROLASE Ligands: GOL, TUU |
 |
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2023-05-24 Classification: HYDROLASE Ligands: GOL, TV6 |
 |
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-05-24 Classification: HYDROLASE Ligands: TVI |
 |
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2023-05-24 Classification: HYDROLASE Ligands: GOL, TVO |
 |
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2023-05-24 Classification: HYDROLASE Ligands: GOL, TW0 |
 |
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2023-05-24 Classification: HYDROLASE Ligands: GOL, TWX |
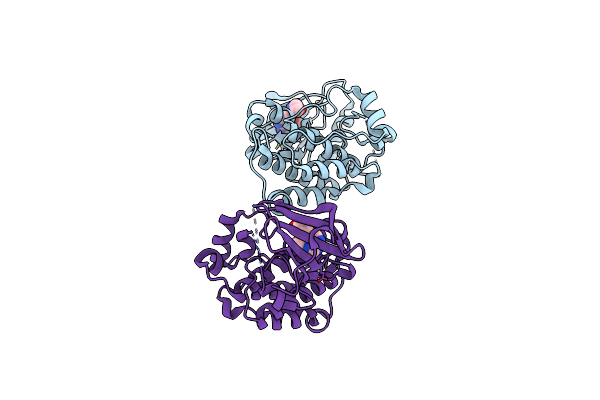 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-10-23 Classification: TRANSFERASE Ligands: GUW |
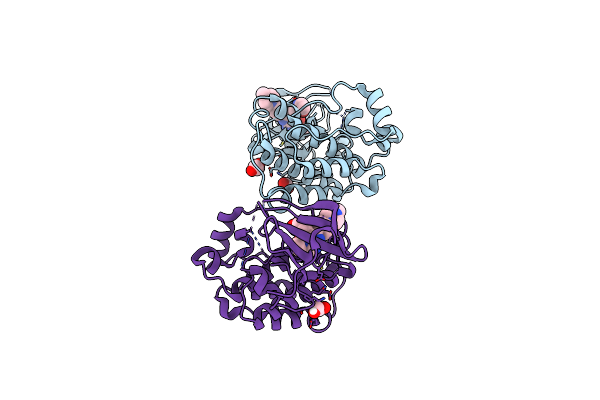 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-10-23 Classification: TRANSFERASE Ligands: GUT, EDO |
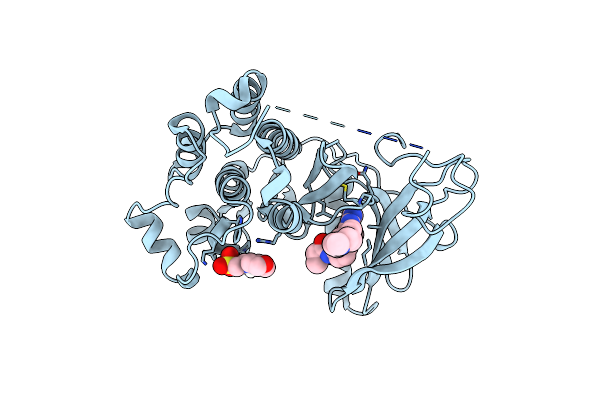 |
Crystal Structure Of Egfr-T790M/C797S In Complex With Covalent Pyrrolopyrimidine 19G
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-10-16 Classification: TRANSFERASE Ligands: L0Q, MES |
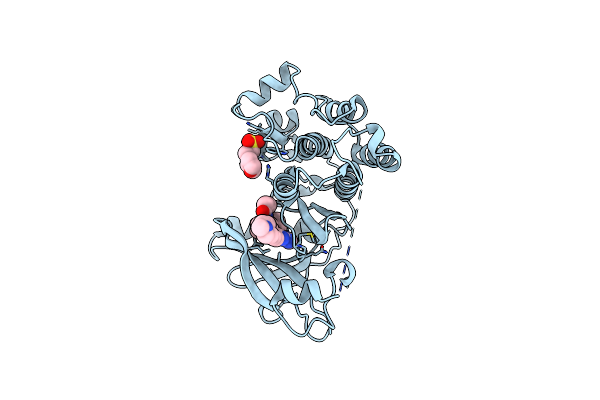 |
Crystal Structure Of Egfr-T790M/C797S In Complex With Covalent Pyrrolopyrimidine 19H
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-10-16 Classification: TRANSFERASE Ligands: MES, L0N |
 |
Crystal Structure Of Phenazine Biosynthesis Protein Phza/B From Burkholderia Cepacia R18194, Apo Form
Organism: Burkholderia sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-12-30 Classification: BIOSYNTHETIC PROTEIN Ligands: ACT |
 |
Crystal Structure Of Phenazine Biosynthesis Protein Phza/B From Burkholderia Cepacia R18194, Complex With 2-(Cyclohexylamino)Benzoic Acid
Organism: Burkholderia sp.
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-12-30 Classification: BIOSYNTHETIC PROTEIN Ligands: 3B4, AZI, ACT |
 |
Crystal Structure Of Phenazine Biosynthesis Protein Phza/B From Burkholderia Cepacia R18194, Dhha Complex
Organism: Burkholderia sp.
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2008-12-30 Classification: BIOSYNTHETIC PROTEIN Ligands: ACT, HHA |
 |
Crystal Structure Of Phza/B From Burkholderia Cepacia R18194 In Complex With (R)-3-Oxocyclohexanecarboxylic Acid
Organism: Burkholderia sp.
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2008-12-30 Classification: BIOSYNTHETIC PROTEIN Ligands: 3OC |
 |
Crystal Structure Of Phza/B From Burkholderia Cepacia R18194 Crystallized In C2221
Organism: Burkholderia sp.
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-12-30 Classification: BIOSYNTHETIC PROTEIN |

