Search Count: 18
 |
Organism: Rhynchosporium secalis
Method: SOLUTION NMR Release Date: 2003-11-11 Classification: TOXIN |
 |
Structure Of The Pyrimidine-Rich Internal Loop In The Y-Domain Of Poliovirus 3'Utr
|
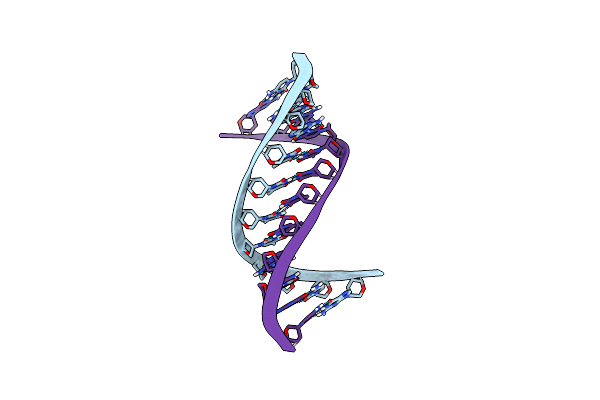 |
Solution Structure Of A Hexitol Nucleic Acid Duplex With Four Consecutive T:T Base Pairs
|
 |
Solution Structure Of The Single-Stranded Dna-Binding Cold Shock Domain (Csd) Of Human Y-Box Protein 1 (Yb1) Determined By Nmr (10 Lowest Energy Structures)
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2002-02-21 Classification: TRANSLATION FACTOR |
 |
Solution Structure Of The Pseudoknot Of Srv-1 Rna, Involved In Ribosomal Frameshifting
|
 |
Structure Of The Ribozyme Substrate Hairpin Of Neurospora Vs Rna. A Close Look At The Cleavage Site
|
 |
Structure And Mechanism Of Formation Of The H-Y5 Isomer Of An Intramolecular Dna Triple Helix.
|
 |
Solution Nmr Structures Of The Major Coat Protein Of Filamentous Bacteriophage M13 Solubilized In Dodecylphosphocholine Micelles, 25 Lowest Energy Structures
Organism: Enterobacteria phage m13
Method: SOLUTION NMR Release Date: 1998-11-11 Classification: VIRAL PROTEIN |
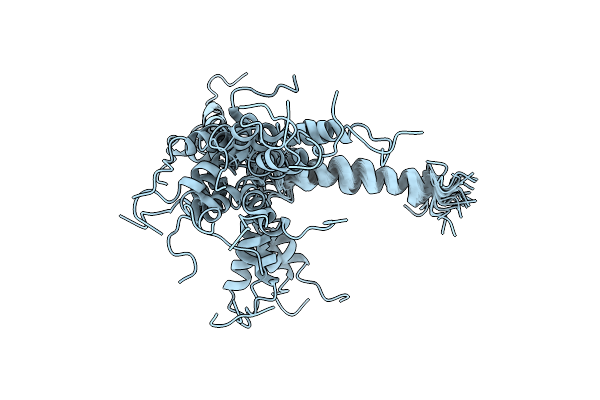 |
Solution Nmr Structures Of The Major Coat Protein Of Filamentous Bacteriophage M13 Solubilized In Sodium Dodecyl Sulphate Micelles, 25 Lowest Energy Structures
Organism: Enterobacteria phage m13
Method: SOLUTION NMR Release Date: 1998-11-11 Classification: VIRAL PROTEIN |
 |
Structure Of The 3' Hairpin Of The Tymv Pseudoknot: Preformation In Rna Folding
|
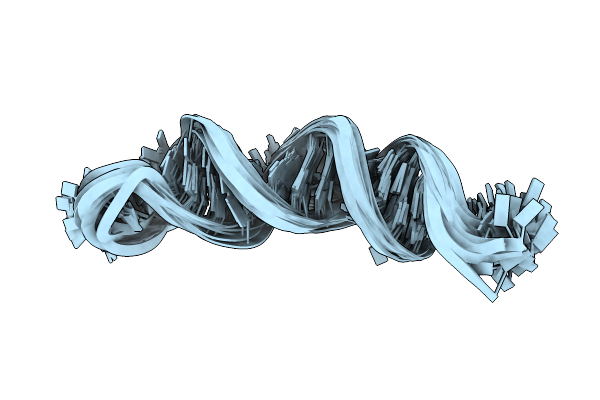 |
Nmr Structure Of A Classical Pseudoknot: Interplay Of Single-And Double-Stranded Rna, 24 Structures
Organism: Turnip yellow mosaic virus
Method: SOLUTION NMR Release Date: 1998-05-27 Classification: RNA |
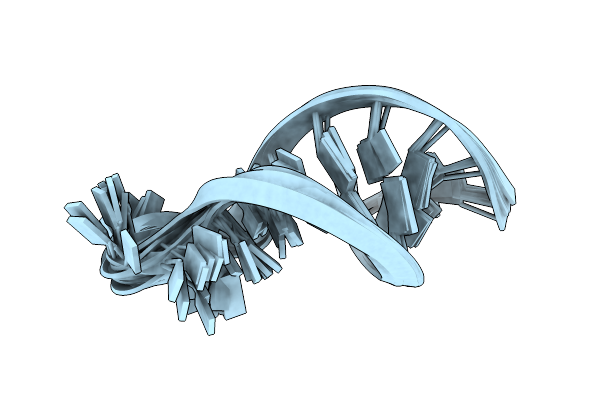 |
The Structure Of The Isolated, Central Hairpin Of The Hdv Antigenomic Ribozyme, Nmr, 10 Structures
|
 |
Structural Features Of The Dna Hairpin D(Atcctagttataggat): The Formation Of A G-A Base Pair In The Loop, Nmr, 10 Structures
|
 |
Solution Nmr Structure Of The Single-Stranded Dna Binding Protein Of The Filamentous Pseudomonas Phage Pf3, Minimized Average Structure
Organism: Pseudomonas phage pf3
Method: SOLUTION NMR Release Date: 1997-02-12 Classification: DNA BINDING PROTEIN |
 |
Refined Solution Structure Of The Tyr 41--> His Mutant Of The M13 Gene V Protein. A Comparison With The Crystal Structure
Organism: Enterobacteria phage m13
Method: SOLUTION NMR Release Date: 1995-10-15 Classification: DNA-BINDING (VIRAL) |
 |
Refined Solution Structure Of The Tyr 41--> His Mutant Of The M13 Gene V Protein. A Comparison With The Crystal Structure
Organism: Enterobacteria phage m13
Method: SOLUTION NMR Release Date: 1995-10-15 Classification: DNA-BINDING (VIRAL) |
 |
Crystal Structures Of Y41H And Y41F Mutants Of Gene V Protein From Ff Phage Suggest Possible Protein-Protein Interactions In Gvp-Ssdna Complex
Organism: Enterobacteria phage f1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1994-06-22 Classification: DNA BINDING PROTEIN |
 |
Crystal Structures Of Y41H And Y41F Mutants Of Gene V Protein From Ff Phage Suggest Possible Protein-Protein Interactions In Gvp-Ssdna Complex
Organism: Enterobacteria phage f1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1994-06-22 Classification: DNA BINDING PROTEIN |

