Search Count: 14
 |
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MG, MPD |
 |
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MPD, MG |
 |
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MG, MPD |
 |
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MPD, MG |
 |
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MG, MPD |
 |
Organism: Thermococcus kodakarensis kod1
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-11-02 Classification: HYDROLASE |
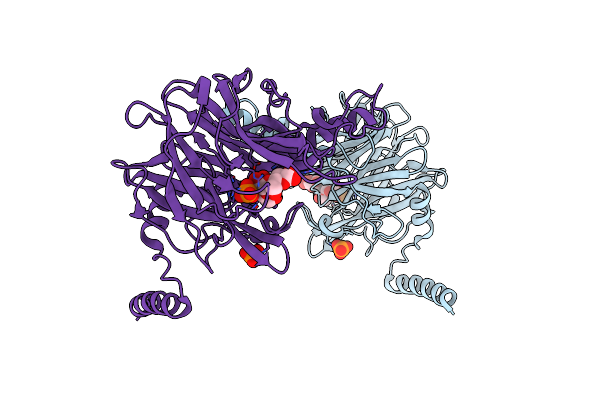 |
Crystal Structure Of 4-O-Beta-D-Mannosyl-D-Glucose Phosphorylase Mgp Complexed With Man-Glc+Po4
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-09-04 Classification: TRANSFERASE Ligands: PO4 |
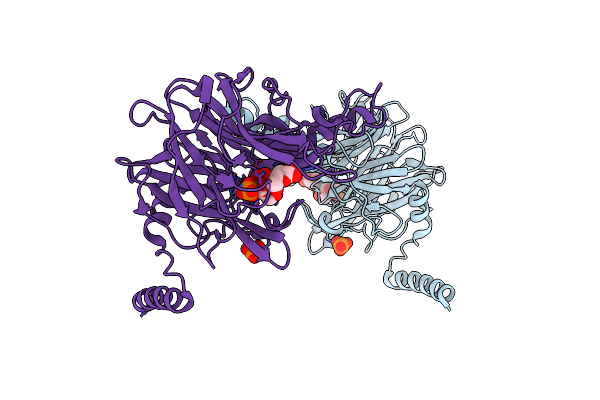 |
Crystal Structure Of 4-O-Beta-D-Mannosyl-D-Glucose Phosphorylase Mgp Complexed With Man+Glc
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-09-04 Classification: TRANSFERASE Ligands: PO4, BGC, BMA |
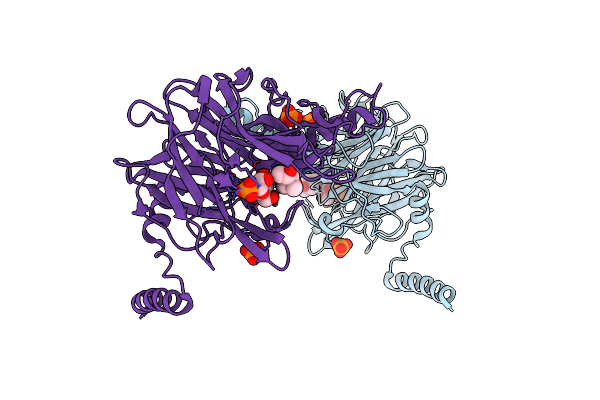 |
Crystal Structure Of 4-O-Beta-D-Mannosyl-D-Glucose Phosphorylase Mgp Complexed With M1P
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-09-04 Classification: TRANSFERASE Ligands: PO4, MPD, M1P |
 |
Crystal Structure Of 4-O-Beta-D-Mannosyl-D-Glucose Phosphorylase Mgp Complexed With Po4
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-09-04 Classification: TRANSFERASE Ligands: PO4, MPD |
 |
Organism: Aspergillus fumigatus af293
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-05-15 Classification: HYDROLASE INHIBITOR |
 |
Organism: Aspergillus fumigatus af293
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-05-15 Classification: HYDROLASE INHIBITOR |
 |
Crystal Structure Analysis Of Neutral Ceramidase From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2009-03-03 Classification: HYDROLASE Ligands: ZN, MG, FMT, PLM, GOL |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-02-03 Classification: HYDROLASE Ligands: ZN, 2ED, FMT, MG, DMS |

